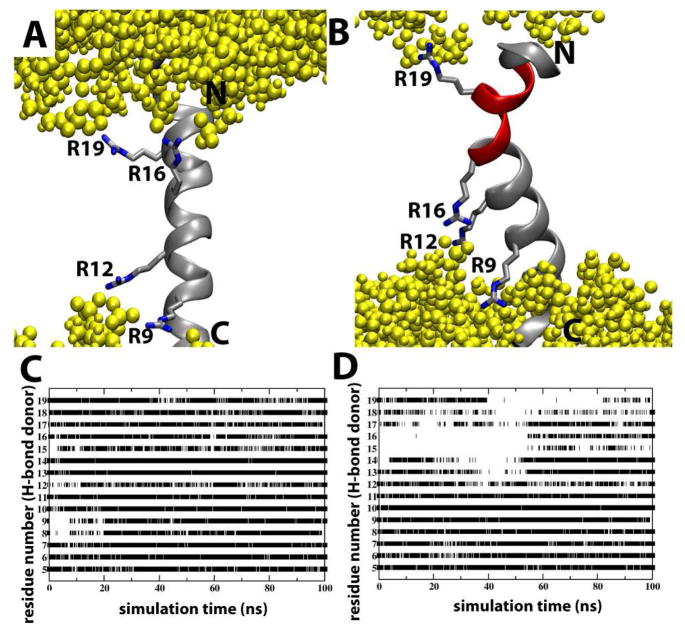
Figure 2.
The effect of Arg rotamers on S4mut conformation in POPC bilayers. (A) A representative simulation snapshot of the S4mut helix with a 2-up-2-down distribution of its Arg residues (R9, R12, R16 and R19). Phospholipid headgroups are colored yellow. (B) A representative simulation snapshot of the S4mut helix with a 1-up-3-down distribution of its Arg residues (R9, R12, R16 and R19). The snapshot shows the severely kinked region (red) of S4mut. (C) The presence of backbone H-bonds in S4mut from an H-bond donor at position i to an H-bond acceptor at position i+4 for the 1-up-3-down Arg distribution and (D) the 2-up-2-down Arg distribution. The flanking -GPGG- regions were left out of this analysis. The criteria for H-bonds in this analysis were defined by cut-off values; 3.5 Å for the distance and 40 degrees for the angle.