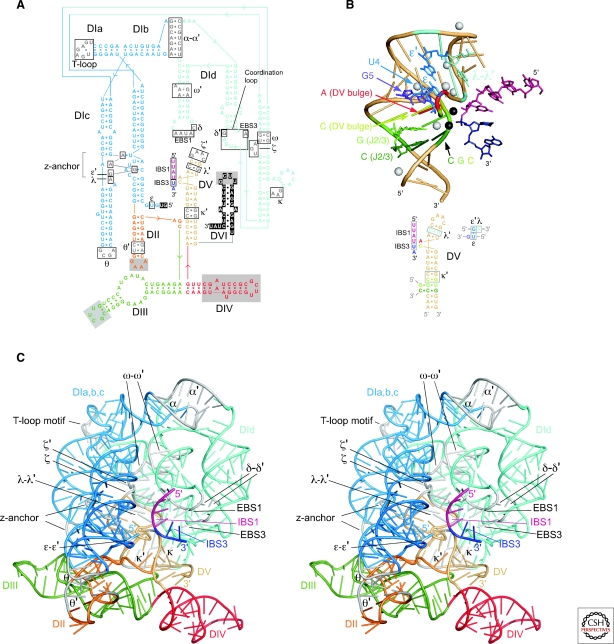
Figure 5.
Crystal structure of the Oceanobacillus iheyensis group IIC intron. (A) Sequence and secondary structure of the crystallized RNA. Boxes indicate motifs involved in tertiary interactions. Solid gray boxes in DII, DIII, and DIV indicate regions deleted from the crystallization construct and replaced with sequences not present in the wild-type intron. Nucleotide residues in DI and DVI shown as white letters on a black background are not visible in the crystal structure. (B) Structure of the active-site region, with a corresponding color-coded secondary structure below. DV is a beige tube helix, with a bound RNA modeled as the 5′ and 3′ exons (pink and indigo, respectively; Toor et al. 2008b; Toor et al. 2010). The triple interactions in the triple helix stack between the CGC triad, the CG of J2/3, and the C of the AC bulge are shown in dark green, green and yellow-green. The three-base stack consisting of the A of the AC bulge, G5, and U4 of the ε–ε′ interaction are in red, purple and blue, respectively, while the λ–λ′ interaction is cyan. Metal ions bound to DV in the crystal are indicated by spheres, with black spheres representing the proposed active-site Mg++ ions, which were identified by binding of Yb3+ in the crystal derivatives. (C) X-ray crystal structure. A stereoview is shown, with domains colored as in (A) and regions involved in tertiary interactions colored gray (Toor et al. 2008a,b). Note added in proof: The conserved single base pair in the κ stem-loop, which we noted was missing in the original structure (Toor et al. 2008a), is present in the recently corrected and refined structure (Toor et al. 2010).