Abstract
Organisms show striking differences in genome structure; however, the functional implications and fundamental forces that govern these differences remain obscure. The intron–exon organization of nuclear genes is involved in a particularly large variety of structures and functional roles. We performed a 22-species study of Meis/hth genes, intron-rich homeodomain-containing transcription factors involved in a wide range of developmental processes. Our study revealed three surprising results that suggest important and very different functions for Meis intron–exon structures. First, we find unexpected conservation across species of intron positions and lengths along most of the Meis locus. This contrasts with the high degree of structural divergence found in genome-wide studies and may attest to conserved regulatory elements residing within these conserved introns. Second, we find very different evolutionary histories for the 5′ and 3′ regions of the gene. The 5′-most 10 exons, which encode the highly conserved Meis domain and homeodomain, show striking conservation. By contrast, the 3′ of the gene, which encodes several domains implicated in transcriptional activation and response to cell signaling, shows a remarkably active evolutionary history, with diverse isoforms and frequent creation and loss of new exons and splice sites. This region-specific diversity suggests evolutionary “tinkering,” with alternative splicing allowing for more subtle regulation of protein function. Third, we find a large number of cases of convergent evolution in the 3′ region, including 1) parallel losses of ancestral coding sequence, 2) parallel gains of external and internal splice sites, and 3) recurrent truncation of C-terminal coding regions. These results attest to the importance of locus-specific splicing functions in differences in structural evolution across genes, as well as to commonalities of forces shaping the evolution of individual genes along different lineages.
Keywords: intron–exon structures, alternative splicing, homeobox transcription factors, convergent evolution
Introduction
Intron–exon structures are highly variable both between and within species. Within metazoans, some species such as humans have an average of ∼9 introns per gene, whereas others, such as flies, have nearly three times less (Roy and Irimia 2009b). A large number of genome-wide interspecies comparisons of intron–exon structures have revealed the history of change and stasis in intron–exon structures underlying these differences. Modern differences largely reflect orders-of-magnitude differences in the rates of intron creation and loss between species (Roy and Penny 2006). At one extreme, orthologous genes from deeply diverged species including vertebrates, the cnidarian Nematostella, and the placozoan Trichoplax have nearly identical intron–exon structures within conserved coding regions, indicating a striking dearth of intron creation and loss changes across hundreds of millions of years (Roy et al. 2003; Coulombe-Huntington and Majewski 2007a; Putnam et al. 2007; Srivastava et al. 2008). At the other extreme, intron positions in lineages such as urochordates and Caenorhabditis nematodes only rarely correspond to intron positions in other lineages, indicating wholesale intron loss and gain (Seo et al. 2001; Rogozin et al. 2003; Edvardsen et al. 2004; Coulombe-Huntington and Majewski 2007b; Putnam et al. 2008).
While powerful for understanding general evolutionary trends, such studies may overlook differences in evolutionary mode between different genes or different introns within the same species. Intron–exon structures vary dramatically across genes: within humans, intron numbers range from hundreds of intronless genes to the 363-exon TITIN gene (Bang et al. 2001), and intron lengths span four orders-of-magnitude (from ∼100 bp to ∼1 Mbp). It is known that splicing encodes a large number of locus-specific functions (production of specific alternative transcripts, regulation of specific genes by production of sterile transcripts by splicing, etc. [e.g., Schmucker et al. 2000; Irimia et al. 2010]); as such, intron function and level of dispensability is likely to vary considerably across genes and introns within the same species. Systematic evolutionary differences have also been observed, often related to transcript position. For instance, in some species, the first (5′-most) intron within a coding sequence tends to be longer, and to exhibit more interspecific sequence conservation, consistent with greater frequencies of functional elements (Bergman and Kreitman 2001; Marais et al. 2005; Hughes et al. 2008). Striking differences in the incidence and lengths of introns are also observed between translated and untranslated regions of genes (Hong et al. 2006; Scofield et al. 2007; Hughes et al. 2008), also suggesting different evolutionary dynamics in different classes of introns. However, genome-wide studies tend to average across introns of different modes and levels of functionality, perhaps inaccurately sketching a portrait of intron evolution as a largely stochastic and random process.
Here, we employ an alternative approach, using many-species studies of an individual gene family to try to discern commonalities of evolution across species and differences between introns within the same genome. We studied myeloid ecotropic viral integration site homologue (Meis) genes (Moskow et al. 1995, called homothorax [hth] in Drosophila [Rieckhof et al. 1997]). In contrast to most homeobox genes, which contain one or no introns, Meis genes contain 10 or 11 introns in most metazoans. Meis are deeply conserved homeodomain-containing transcription factors of the TALE (three-amino acid loop extension) superclass, involved in a wide variety biological processes, ranging from hematopoiesis (Hisa et al. 2004; Azcoitia et al. 2005) to limb development and regeneration (Mercader et al. 1999; Mercader et al. 2005). Meis1 and Meis2 have overlapping but distinct dynamic expression domains in the developing central nervous system, related to patterning of the developing telencephalon (Toresson et al. 2000), pretectum (Ferran et al. 2007), and hindbrain (Dibner et al. 2001; Choe et al. 2002; Wassef et al. 2008). In Drosophila, hth has also been implicated in several biological processes, some of them in common with vertebrates (Pai et al. 1998; Mercader et al. 1999).
Most vertebrates contain three paralogs of Meis (Nakamura et al. 1996; Sánchez-Guardado et al. 2011), dating to the two rounds of whole-genome duplication (WGD) at the base of vertebrates (Dehal and Boore 2005; Putnam et al. 2008). Adding to MEIS protein diversity, Meis genes have been shown to be alternatively spliced. For instance, exon “12a” of the vertebrate Meis1 gene is alternatively spliced: the Meis1A isoform contains exon 12a (fig. 1A), but the Meis1B isoform does not, leading to an alternative C-terminus, encoded by the downstream exon 12b, and to higher transcriptional activator capacities than both Meis1A- and the Meis-related pknox1 gene, especially in response to protein kinase A (PKA) and TrichostatinA (TSA) (Maeda et al. 2001; Huang et al. 2005). Alternative splicing (AS) of exons homologous to 12a, as well as other AS events, have been reported for the Meis2 and Meis3 genes in vertebrates (Oulad-Abdelghani et al. 1997; Yang et al. 2000; Williams et al. 2005; Shim et al. 2007; Hyman-Walsh et al. 2010; Sánchez-Guardado et al. 2011).
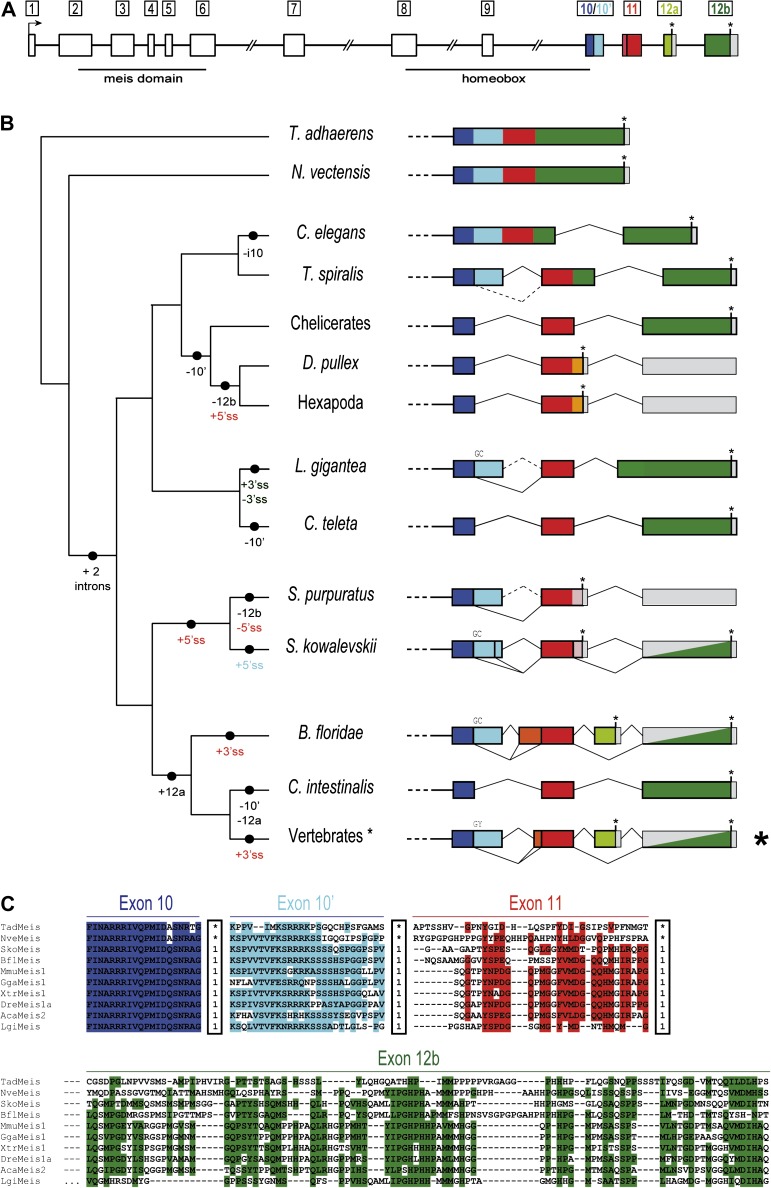
FIG. 1.
Evolution of the intron–exon structures of the C-termini of Meis/hth. (A) Schematic representation of the intron–exon structure of a prototypical vertebrate Meis gene. The conserved long size of introns 6–9 are indicated by a double slash. Homologous coding regions (exons) in the 3′ are indicated by colors: 10 (dark blue), 10′ (light blue), 11 (red), 12a (light green), and 12b (dark green). (B) Diversity of intron–exon structures of exons 10–12b in metazoans. The different genomic gains (+) and losses (−) of regions or splice sites (5′ splice site (SS) or 3′ ss, colored according to the exon), assuming parsimony, are indicated in the branches of the schematic tree on the left-hand side. Solid vertical bars between colors represent a conserved 5′ ss, and GC 5′ ss are indicated above each line. Asterisks represent termination codons and gray blocks indicate UTR exons. Split gray/colored boxes indicate regions that are either translated or 3′ UTR depending on splice form. (C) Sequence alignment for some representative bilaterians and the two non-bilaterians showing sequence conservation at each exon. Within the boxes, “1” indicates a phase 1 intron, and an asterisk represents absence of an intron at that position. Highlighted positions correspond to 60% of similar amino acid types across studied genes, as generated by BioEdit.
Here we investigate the evolution of intron–exon structures and AS of Meis genes across metazoans. We find very different evolutionary histories for the 5′ and 3′ regions of the gene. Intron–exon structures of the 5′-most region, corresponding to the first ∼1,000 nt of the coding sequence, are highly similar across species, with the positions and relative sizes of the first 9 intron positions being highly conserved across studied species. Unexpectedly, this conservation extends to metazoan groups with intron–exon structures that are generally very divergent, such as flies, nematodes and tunicates, suggesting functional constraints opposing intron loss. On the other hand, the C-terminal coding regions exhibit a complex and surprising history marked by creation and loss of introns and exons, gain and loss of AS of various gene regions, and a remarkable variety of cases of parallel evolution at the levels of genome, gene transcripts, and gene function. These differences in the evolution of intron–exon structures and splicing across Meis genes are likely to reflect, at least in part, qualitatively different protein and regulatory functions encoded by different genic regions. These results underscore the utility of many-species studies for understanding the functional genomics of introns and splicing.
Materials and Methods
Genome Sources and Gene Annotation
We used the following genome sequence assemblies and expression data (expressed sequence tags [ESTs]) from the following sources: Trichoplax adhaerens Grell-BS-1999 v1.0 (Srivastava et al. 2008), N. vectensis v1.0 (Putnam et al. 2007), Branchiostoma floridae v1.0 (Putnam et al. 2008), Ciona intestinalis v2.0 and v1.0 (Dehal et al. 2002), Takifugu rubripes v4.0, Xenopus tropicalis v4.1 (Hellsten et al. 2010), Daphnia pulex v1.0, Helobdella robusta v1.0, Lottia gigantea v1.0 and Capitella teleta v1.0, at DOE Joint Genome Institute (JGI) Web page (http://genome.jgi-psf.org/euk_home.html), and of Strongylocentrotus purpuratus Build 2.1 (Sea Urchin Genome Sequencing Consortium et al. 2006), Apis mellifera Amel_4.0, Tribolium castaneum Build 2.1 (Tribolium Genome Sequencing Consortium et al. 2008), Drosophila melanogaster Build Fb5.3 (Adams et al. 2000), Danio rerio Zv8, Gallus gallus v2.1 (Chicken Genome Sequencing Consortium 2004), Anolis carolinensis AnoCar1.0, Homo sapiens Build GRCh37 (Lander et al. 2001; Venter et al. 2001), Mus musculus Build 37.1 (Waterston et al. 2002), and Acyrtosiphon pisum Build 1.1, at the NCBI Web page (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) and/or Ensembl Web page (http://www.ensembl.org), Trichinella spiralis at the NCBI Web page for unfinished eukaryotic genomes (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi?organism=eukaryotes), Brugia malayi BMA1 (Ghedin et al. 2007) at TIGR Web page (http://blast.jcvi.org/er-blast/index.cgi?project=bma1), Caenorhabditis elegans WS213 (Caenorhabditis elegans Sequencing Consortium 1998) at WormBase (http://www.wormbase.org), and Saccoglossus kowalevskii 09 December 2008 scaffolds at HGSC Baylor College of Medicine Web page (http://blast.hgsc.bcm.tmc.edu/blast.hgsc?organism=20). Additional sequences from arthropods without available genome resources (those included in supplementary fig. S1) were retrieved through TBlastN searches against the nucleotide collection database at the NCBI Web page. In more poorly annotated genomes, Meis candidates were searched by TBlastN and gene annotation was then performed by downloading the whole associated genomic region and identifying each exon by mapping available expression data and/or by similarity of sequence using ClustalW and Blast2seq. Available automatic gene predictions were also used. Combining the different sources of data, most exon boundaries could be determined unambiguously (supplementary table S1). Introns and exons were named following the vertebrate nomenclature (exons 12 and 13 were named 12a and 12b [Sánchez-Guardado et al. 2011] and insect-specific exons between ancestral exons 7 and 8 were not counted [supplementary fig. S1]). Intron–exon structures of the 5' untranslated regions (UTRs) are not described here due to the lack of expression data for most species and difficulty to assess intron position conservation in noncoding sequences over long phylogenetic ranges.
Median, average and average excluding the top 5% intron lengths and intergenic distances (supplementary table S2) were calculated for each genome using custom Perl scripts on GTF (Ensemble), GFF (JGI), or GBK (NCBI) files or obtained from Irimia, Maeso, and Garcia-Fernandez (2008).
Phylogenetic Analyses
Meis/hth protein sequences from multiple species were aligned using MAFFT (Katoh et al. 2002, 2005) as implemented in Jalview 2.4 (Waterhouse et al. 2009), and the alignments were manually curated by using information on intron positions (Irimia and Roy 2008). Two different phylogenetic analyses were performed. First, to establish orthology of all studied Meis/hth genes, we used an alignment containing only the highly conserved Meis and Homeobox domains and including several Meis-related pknox proteins as outgroups (supplementary fig. S2A). Second, to allow confident assignment of paralogy relationships within vertebrates, the number of positions included in the alignment was increased using the whole protein sequence (except exon 1 and the alternatively spliced 3′ regions [exons 10′ to 12b]), and fast-evolving species were excluded (supplementary fig. S2B). Phylogenetic trees were generated by the Bayesian method with MrBayes 3.1.2 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003) using two independent runs (each with four chains). Model selection using ProtTest (Drummond and Strimmer 2001; Guindon and Gascuel 2003; Abascal et al. 2005), convergence determination, burn-in, and consensus tree calculations were done as previously described (D'Aniello et al. 2008).
cDNA Samples and Reverse Transcription–Polymerase Chain Reaction of Alternative Splicing Events
RNA from adult and/or embryonic vertebrate (D. rerio, X. tropicalis, A. carolinensis, G. gallus, and M. musculus) tissues and different amphioxus (B. lanceolatum) developmental stages was extracted using RNeasy Mini Kit (Qiagen), and retrotranscriptions were done using SuperScript III Reverse Transcriptase (Invitrogen), according to manufacturer. One A. carolinensis adult animal was bought in a local pet shop. All animals were sacrificed following standard and ethically approved procedures by the European Union and the Spanish government for laboratory animals.
For reverse transcription–polymerase chain reaction (RT-PCR) analyses, we designed two sets of primers for each gene in each studied species (S. purpuratus, B. lanceolatum, D. rerio, X. tropicalis, A. carolinensis, G. gallus, and M. musculus). The first set spans exons 10, 10′, and 11 and the second one exons 11, 12a (when present), and12b, to yield all isoforms of the 3′ region present in the studied set of tissues. All primer sequences are provided in supplementary table S3. RT-PCR were done trying to minimize the number of cycles and at least 3′ of elongation to diminish the PCR bias for short isoforms (Rukov et al. 2007), except for those probing exon 10′ inclusion (supplementary fig. S3), for which we used 36 cycles in each of two rounds of amplification.
Results
Meis Gene Complements in Metazoans
We studied 7 vertebrate and 15 invertebrate genomes, spanning all major metazoan clades (deuterostomes, protostomes, lophotrochozoans, and non-bilaterians). In most studied invertebrates, we found only 1 Meis/hth ortholog, including the basal branching non-bilaterians T. adhaerens and N. vectensis. However, we found four Meis genes in the lophotrochozoan H. robusta; in addition, two paralogs have been described in two distantly related spiders (Prpic et al. 2003; Pechmann and Prpic 2009), Cupiennius salei and Acanthoscurria geniculata, for which full genome sequences are not yet available. Among vertebrates, Meis complement ranged from two paralogs in birds (Sánchez-Guardado et al. 2011) to five in zebrafish (dating to the extra round of WGD that occurred at the base of teleosts), with three genes in most tetrapods. Phylogenetic analysis using Bayesian inference strongly supports the orthology of all identified genes (supplementary fig. S2).
High Level of Conservation of 5′ Intron–Exon Structures of Meis across Metazoans
To compare intron-exon structures of Meis across animal lineages, we mapped intron positions onto alignments of translated coding sequences. We found very different general patterns for the 5′ and 3′ portions of the gene (fig. 1A). The first 9 intron positions and phases were conserved in all studied metazoans, with the two exceptions of the loss of intron 4 in C. elegans and of intron 3 in a divergent paralog in the leech H. robusta and intron gain events splitting exon 6 independently in B. malayi and another paralog of H. robusta. This extreme conservation is in striking contrast to the general patterns found in genome-wide studies, in which intron positions in a variety of lineages, notably arthropods, nematodes, and tunicates, show very little correspondence, attesting to large amounts of intron loss and gain (Logsdon 2004; Rogozin et al. 2003; Edvardsen et al. 2004; Putnam et al. 2008). The finding of widespread intron position correspondence in the 5′ regions of Meis genes thus suggests that locus-specific forces opposing loss of ancestral introns and gain of new ones are acting across a wide variety of metazoan lineages.
In addition, we found that the relative sizes of introns are widely conserved across species. In nearly all studied species, introns 6–9 are the longest (P < 0.0001 in a Kolmogorov–Smirnov comparison between introns 6 and 9 vs. the rest), with sizes usually 10–30 times larger than the species average intron length, reaching ∼100 times as long as the average in some extreme cases (table 1). This pattern is observed both in vertebrates and invertebrates and in large and compact genomes. For instance, out of only ∼500 introns longer than 10 Kb in the compact genome of the pufferfish T. rubripes (Aparicio et al. 2002), 6 are found in 2 Meis paralogs. Long introns are often associated with regulatory signals contained within intronic sequences; a regulatory role for these long introns could explain the lack of intron loss in diverse lineages (see below).
Table 1.
Length of Each Intron and Species Median, Average, and Average Excluding the Longest 5% Introns
| Median | Average | Average − 5% | Intron 1 | Intron 2 | Intron 3 | Intron 4 | Intron 5 | Intron 6 | Intron 7 | Intron 8 | Intron 9 | Intron 10 | Intron 11 | Intron 12 | Mean | |
| H. sapiens Meis1 | 1,419 | 5,787 | 2,792 | 1,868 | 1,879 | 577 | 801 | 1,437 | 21,060 | 47,928 | 35,648 | 19,433 | 1,155 | 293 | 2,100 | 11,182 |
| H. sapiens Meis2 | 1,255 | 1,536 | 674 | 986 | 796 | 9,695 | 46,811 | 86,413 | 53,637 | 1,366 | 338 | 2,257 | 17,147 | |||
| H. sapiens Meis3 | 1,719 | 214 | 101 | 1,550 | 109 | 5,169 | 188 | 1,961 | 107 | 361 | — | 2,906 | 13,08 | |||
| M. musculus Meis1 | 1,290 | 4738 | 2,260 | 1,861 | 1,741 | 567 | 771 | 1,394 | 22,862 | 46,463 | 35,893 | 20,037 | 1,116 | 304 | 2,065 | 11,256 |
| M. musculus Meis2 | 671 | 1,595 | 719 | 1,116 | 733 | 9,432 | 48,807 | 78,382 | 52,868 | 1,411 | 364 | 2,292 | 16,533 | |||
| M. musculus Meis3 | 1,824 | 135 | 92 | 733 | 155 | 2,940 | 167 | 1,322 | 170 | 538 | 729 | 539 | 779 | |||
| G. gallus Meis1 | 806 | 2,616 | 1,332 | ? | ? | ? | ? | >849 | >6,084 | 25,232 | 25,453 | 16,211 | 1,271 | 188 | 1,919 | 11,712 |
| G. gallus Meis2 | ? | >967 | 222 | 1,734 | 791 | 10,760 | 34,904 | 59,559 | 54,586 | 1,439 | 422 | 3,103 | 16,752 | |||
| T. rubripes Meis1.1 | 147 | 568 | 315 | 1,491 | 999 | 219 | 425 | 1,010 | 7,736 | 15,920 | 13,058 | 10,547 | 1,832 | 677 | — | 4,901 |
| T. rubripes Meis1.2 | 1,061 | 388 | 243 | 78 | 70 | 114 | 1,051 | 1,388 | 1,016 | 373 | 145 | — | 539 | |||
| T. rubripes Meis2 | 225 | 1,604 | 233 | 326 | 371 | 7,877 | 18,177 | 18,491 | 14,252 | 912 | 368 | 3,260 | 5,508 | |||
| T. rubripes Meis3 | 2,027 | 301 | 93 | 87 | 84 | 79 | 138 | 371 | 817 | 87 | — | 90 | 379 | |||
| C. intestinalis | 333 | 545 | 365 | 2,488 | 4,548 | 1,349 | 1,357 | 363 | 4,275 | 4,147 | 2,197 | 6,648 | 101 | — | 242 | 2,520 |
| B. floridae | 730 | 1,460 | 973 | 177 | 412 | 633 | 701 | 1,231 | 3,513 | 9,733 | 9,140 | 9,863 | 2,580 | 503 | 2,552 | 3,420 |
| S. kowalevskii | n.d. | n.d. | n.d. | 274 | 453 | 697 | 1,289 | 1,943 | 5,390 | 9,712 | 6,089 | 11,058 | 1,815 | — | — | 3,872 |
| S. purpuratus | 748 | 1,624 | 1,015 | 297 | 850 | 1,965 | 901 | 10,371 | 36,100 | 29,274 | 3,725 | 11,805 | 6,788 | — | — | 10,208 |
| D. melanogaster | 74 | 1,123 | 394 | 7,616 | 2,154 | 9,190 | 2,091 | 5,480 | 23,710 | 40,688 | 6,727 | 2,450 | 221 | — | — | 10,033 |
| A. mellifera | 120 | 1,177 | 334 | 14,086 | 8,857 | 8,878 | 20,587 | 9,010 | 69,950 | 13,9655 | 63,262 | 73,730 | 21,383 | — | — | 42,940 |
| T. castaneum | 54 | 1,312 | 553 | 4,989 | 9,963 | 14,951 | 1,722 | 3,163 | 5,821 | 28,551 | 3,236 | 6,276 | 629 | — | — | 7,930 |
| D. pulex | 294 | 491 | 333 | 2,689 | 1845 | 4,383 | 1394 | 1,334 | 10,586 | 26,222 | 1,323 | 14,569 | 618 | — | — | 64,96 |
| I. scapularis | n.d. | n.d. | n.d. | ? | 18,062 | 8,679 | 2356 | 17,775 | 63,774 | 10,0420 | 31,473 | 46,591 | 3,458 | — | 2,754 | 29,534 |
| C. elegans | 65 | 302 | 204 | 1,411 | 318 | 47 | — | 558 | 1,122 | 1,128 | 312 | 3,973 | — | 643 | 100 | 961 |
| T. spiralis | n.d. | n.d. | n.d. | 922 | 522 | 57 | 134 | 683 | 669 | 1,037 | 1,436 | 5,423 | 193 | — | 60 | 1,012 |
| L. gigantea | 552 | 965 | 662 | 129 | 536 | 1,608 | 931 | 804 | 17,612 | 17,709 | 10,687 | 18,634 | 1,330 | -- | 182 | 6,378 |
| C. teleta | 299 | 553 | 386 | 1,502 | 169 | 505 | 533 | 2,883 | 5,995 | 13,235 | 2,197 | 5,760 | 124 | -- | 3,361 | 3,297 |
| N. vectensis | 591 | 961 | 723 | ? | 304 | 183 | 249 | 381 | 1,062 | 3,494 | 2,409 | 4,920 | — | — | — | 16,25 |
| T. adhaerens | 278 | 419 | 320 | ? | ? | ? | 2,306 | 7,825 | 3,181 | ? | 267 | 904 | — | — | — | 2,897 |
| Average bilaterians | 495 | 1,662 | 851 | 2,373 | 2,651 | 2,520 | 1,944 | 2,742 | 14,340 | 26,633 | 16,421 | 16,843 | 2,208 | 434 | 1,695 | 8,620 |
Note.—“?” indicates that the size could not be determined, “>”minimum size, and “—” intron absence. For genome-wide data, “n.d.” indicates that statistics could not be determined due to lack of genome-wide annotation. Introns 6–9 are shown in italics to highlight their consistently longer lengths across bilaterians.
Interestingly, the only cases in which introns 6–9 are not long relative to species average occur in vertebrates. This could possibly reflect relaxed constraint on intronic regulatory functions following gene duplication. Consistent with this notion, vertebrate paralogs with short introns show more restricted developmental expression domains than do other vertebrate Meis genes (e.g., Meis3 [Waskiewicz et al. 2001; Ng et al. 2009]). Perhaps relatedly, these same paralogs show reduced intergenic lengths. Whereas Meis genes are often found in large genomic regions with extended intergenic regions across animal phylogeny (supplementary table S2), paralogs with shortened introns also show highly reduced intergenic distances relative to other Meis genes. Together these results suggest general loss of regulatory motifs in noncoding regions following gene duplication.
Complex and Convergent Evolution of Meis 3′ Intron-Exon Structures
In contrast to widespread conservation of intron–exon structures in the 5′ region of the gene, the 3′ of metazoan Meis genes showed a much more volatile evolutionary history (fig. 1B); 3′ intron–exon structures differ between bilaterian and non-bilaterian genes: the entire region is encoded by a single exon in both studied non-bilaterians, N. vectensis and T. adhaerens, but is divided into multiple exons in all studied bilaterians. The orthologous sequence in most bilaterians is divided into three exons: one exon which we call exon 10 + 10′ (see below), exon 11, and exon 12b (often called 13, fig. 1). The simplest explanation for this difference is two intron gains at the base of bilaterians.
The region also shows remarkable diversity within bilaterians (fig. 1B). First, the 10th exon is alternatively spliced in diverse bilaterian lineages, with usage of an alternative splice site within the exon. (We refer to the upstream constitutive region as exon 10 and the downstream alternatively spliced region as 10′.) Interestingly, in many bilaterian lineages, the upstream 5′ splice site is a rare GC (accounting for 46% of splice boundaries at this position in studied genes; indicated in Figures 1 and 2). Whereas exon 10 is present in all transcripts, splicing of 10′ varies widely across groups, with 4 observed patterns: 1) 10′ is included in all available transcripts (nematodes), 2) a significant fraction of the transcripts show 10′ inclusion (amphioxus and hemichordates, based on RT-PCR and/or EST count), 3) despite clear conservation of exon sequence and coding meaning in the genome, inclusion of 10′ occurs at very low levels (some vertebrate genes) or could not be observed at all in either ESTs or RT-PCR experiments (sea urchin, supplementary fig. S3), and 4) the sequence encoding 10′ have been lost from the genome (some vertebrates, C. teleta, C. intestinalis, and arthropods). (Specifically, sequence clearly homologous to exon 10 is found, but the downstream sequence shows no similarity to the 10′ region, indicating loss of coding potential.) Strikingly, the loss of 10′ at the genomic level (case 4, above) has independently occurred at least nine times in the evolution of the studied genes (figs. 1 and 2).
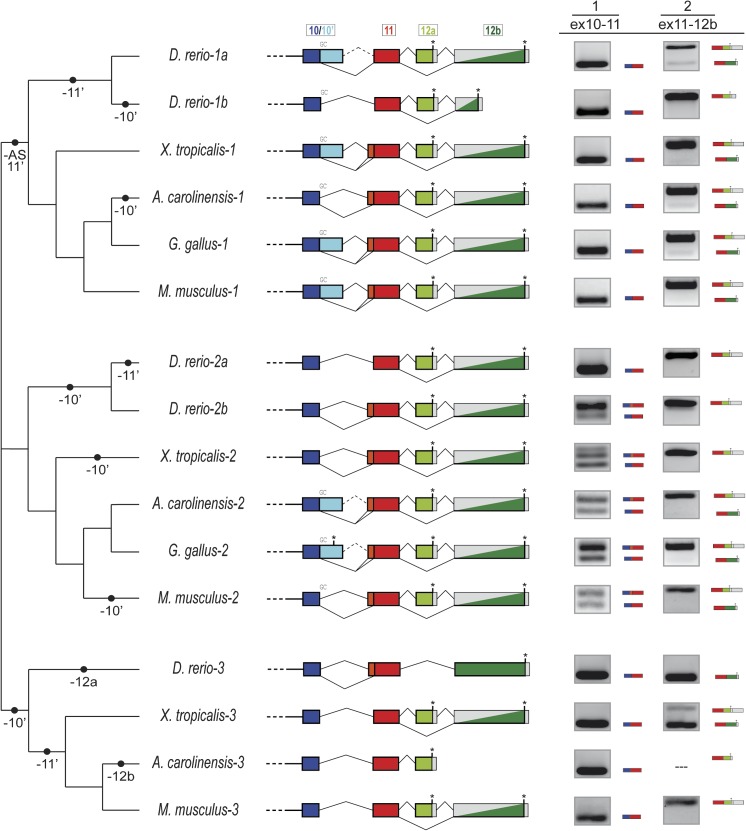
FIG. 2.
Evolution of intron–exon structures and alternative splicing of the 3′ end of vertebrate Meis genes. Diversity of intron–exon structures of exons 10–12b in vertebrates. The different genomic gains (+) and losses (−) of regions, assuming parsimony, are indicated in the branches of the schematic tree on the left-hand side. Solid vertical bars between colors represent a conserved 5′ splice site (SS), and GC 5′ ss are indicated above each line. Asterisks represent termination codons and gray blocks indicate UTR exons. Split gray/colored boxes indicate regions that are either translated or 3′ UTR depending on splice form. RT-PCR results for each event are shown on the right-hand side.
Second, different Meis genes have undergone recurrent truncation at the transcript and genomic level. In each case, truncation has occurred by the introduction of a STOP codon within a novel exonic region upstream of the exon containing the putative ancestral protein terminus (exon 12b; novel and ancestral STOPs are indicated by asterisks in figs. 1 and 2). 1) In some groups, the inferred ancestral situation has been maintained, with the terminal exon constitutively encoding the STOP codon (C. intestinalis, lophotrochozoans, some vertebrates, some ecdysozoans). 2) In chordates, a new alternative STOP-containing exon has arisen by an unknown mechanism (exon 12a, light green in figs. 1 and 2). The new exon is alternatively spliced; its inclusion leads to premature termination, leaving the ancestral exon 12b as 3′ untranslated region (3′ UTR, depicted as half grey). 3) In Ambulacraria (sea urchin and hemichordates), a new downstream splice site has evolved for exon 11, which is alternatively spliced. Use of the new 5′ splice site introduces “extra” downstream sequence (pink), which includes a new STOP codon; as in the case of exon 12a the resulting MEIS protein has a novel C-terminus, and all of exon 12b lies downstream of the STOP codon as 3′ UTR. Interestingly, in sea urchin, the ancestral exon 11 splice site has been lost, implying constitutive use of the new STOP codon and protein truncation. RT-PCR analyses throughout the early development of sea urchin confirmed that only the new terminal isoform is expressed (supplementary fig. S3). 4) Finally, in Pancrustacea (insects and crustaceans [D. pulex]), we found a situation similar to sea urchin: the termination codon is located in a downstream extension of exon 11, and no 12b coding meaning is recognized in the untranslated downstream exon. Importantly, in the last three cases (2–4), the protein sequence, structure, and function of C-terminus of Meis, which harbors the capacities for transcriptional activation and Hox interaction (Yang et al. 2000; Huang et al. 2005; Williams et al. 2005; Hyman-Walsh et al. 2010), are likely to be highly modified relative to the ancestral protein.
Other specific molecular elaborations have also evolved in several lineages (fig. 1B). For instance, the hemichordate S. kowalevskii shows an additional alternative 5′ splice site within the exon 10′ region (i.e., three alternative splice sites for the same exon 10 + 10′), producing an exon with an intermediate length (42 nucleotides less than the entire 10 + 10′ exon). On the other hand, exon 11 in chordates has evolved a 5′ extension of different lengths across species by emergence of an alternative upstream 3′ splice site, resulting in an extended alternative coding sequence (previously described for mammalian Meis2 (Oulad-Abdelghani et al. 1997)). Similarly, in the lophotrochozoan L. gigantea, exon 12b has a constitutive (i.e., not alternative) 5′ extension of ∼35 codons, consistent with loss of the ancestral 3′ splice site and use of a novel upstream site. Finally, nematodes exhibit loss and gain of introns, with loss of the ancestral phase 1 intron between exons 11 and 12b, and gain of a new phase 2 intron at a nearby site in a common ancestor of Trichinella, Brugia, and Caenorhabditis, and subsequent loss of the intron between exons 10′ and 11 in Caenorhabditis (fig. 1B). In stark contrast to this 3′ diversity, only little transcriptional variation was found in the 5' of the gene. AS in insects and vertebrates produce homeodomain-less proteins (which are, indeed, C-terminal truncations) with distinct functions (Yang et al. 2000; Noro et al. 2006); similarly, alternative acceptor site choice within exon 6 in the vertebrate-derived paralog Meis3 results in a protein without a meis domain (Hyman-Walsh et al. 2010). In addition, the annelid C. teleta has a mutually exclusive tandem exon duplication of exon 9 which results in two proteins that differ by only 5 aa substitutions (supplementary table S1), and insects and arthropods harbor 1–3 lineage-specific exons between the ancestral exons 7 and 8 (supplementary fig. S1).
Evolution of Alternative Splicing in Chordate Meis Genes
We next focused on chordates, studying the different AS events of the 3′ regions of Meis genes within 17 genes in 6 chordate species, both in silico and by RT-PCR (fig. 2) and supplementary figs. S4–6. We designed two sets of primers, one spanning exons 10, 10′, and 11 and the other spanning exons 11, (12a), and 12b (see Materials and Methods), and performed RT-PCRs for all Meis genes from six species (amphioxus, zebrafish, X. tropicalis, the anole lizard A. carolinensis, chicken, and mouse), a total of 34 AS events.
For exon 10 + 10′, we found that exon 10′ is included at very low levels in Meis of different vertebrates, only detectable using 10′-specific primers and a high number of PCR cycles for most species and tissues (supplementary fig. S4, see Materials and Methods). This is consistent with observations in human patients (Xiong et al. 2009) and in available ESTs (27/27 and 6/6 ESTs in humans and mouse shown exclusion of exon 10′). Perhaps relatedly, exon 10′ coding meaning has been lost at the genomic level at least 6 times in vertebrate Meis genes, including all Meis3 genes, the Meis2 genes of zebrafish, Xenopus, and mammals, the Meis1 gene of lizard, the Meis1.2 gene of zebrafish; in addition, in-frame STOP codons interrupt this region in chicken Meis2 (fig. 2), suggesting a process of ongoing loss of this region from the gene.
For exon 11, we observed complex patterns for the 5′ extension (orange blocks). This extension with conserved coding meaning (often 21 nt) is found in a wide variety of vertebrate genes, suggesting emergence of an alternative upstream splice site in chordate ancestors. As with exon 10′, the phylogenetic distribution of this alternative splice site is highly punctate, with 4 parallel losses—in zebrafish Meis1.1/.2, zebrafish Meis2.1, tetrapod Meis3 (fig. 2), and C. intestinalis Meis. In addition, although the genomic sequences of both Meis1 and Meis2 genes contain the potential splice site and conserved coding sequence, use of the splice site was only observed in Meis2 genes (fig. 2). The frequency of usage in Meis2 is conserved both across species and development (∼50% in various vertebrates, and throughout different tissues of several vertebrate species [supplementary fig. S5 and Sánchez-Guardado et al. 2011], similar to the pattern in amphioxus [supplementary fig. S6A]).
For exons 12a/12b, exon 12a shows very high levels of inclusion in nearly all paralogs of all vertebrate species; the only exceptions are Xenopus Meis3, which show more moderate levels of inclusion, and zebrafish and human Meis3, which have lost the exon entirely (fig. 2 and supplementary fig. S5). Meis1 paralogs seem to have slightly lower levels of inclusion of exon 12a than Meis2, although the inclusion level is still higher than 90% (fig. 2). The high exon 12a inclusion level was found in a wide range of different tissues in several vertebrate species (supplementary fig. S5), in accordance with previous reports (Azcoitia et al. 2005; Williams et al. 2005; Sánchez-Guardado et al. 2011). Nonetheless, the possibility that a specific cell type in a particular developmental stage show a different splicing pattern cannot be ruled out (e.g. (Oulad-Abdelghani et al. 1997)). However, despite the fact that frequent inclusion of exon 12a may imply only infrequent translation of exon 12b, the ancestral coding meaning of exon 12b has been highly conserved in the vast majority of vertebrate Meis genes; the only exceptions are anole lizard Meis3, which seems to have lost the entire exon, and zebrafish Meis1.2, which has a much shorter sequence. Interestingly, the basal invertebrate chordate amphioxus shows significantly lower levels of exon 12a inclusion (i.e., higher levels of the ancestral isoform [supplementary fig. S6B]).
Discussion
We report the broadest evolutionary comparison of splicing diversity in a homeobox gene family to date. Three aspects of our 22-species comparison of metazoan Meis genes are of particular note: 1) conservation of intron positions and relative sizes across bilaterians, 2) striking differences in diversity and evolutionary patterns between the 5′ and 3′ regions of the gene, suggesting very different functions for introns and splicing for the two regions, and 3) convergent evolution of a variety of features of the alternative transcriptome of the 3′ region, suggesting similar selective forces acting on gene function across widely diverged species. These results show the utility of many-species studies for revealing modes of constraint and innovation acting at individual intronic loci and suggest a general strategy for comparative genomic analysis of splicing function.
Intron–Exon Structures and the Conservation-Implies-Function Paradigm
Comparative genomics has contributed a tremendous amount to our understanding of genome function. Arguably, the most productive paradigm has been “conservation implies function”: in the face of ongoing mutation, only functional genomic features maintained by purifying selection will be retained over long evolutionary times (although for exceptions to this paradigm and a contrasting discussion, see Monroe 2009; Alexander et al. 2010). In the context of base pair substitutions and other small-scale sequence mutations, searches for conservation have utilized baseline mutation rates estimated from rates of changes for various classes of putatively neutral sites (e.g., synonymous or intronic sites) in order to identify slow-evolving and thus putatively functional sequences. While formally applicable to intron–exon structures, this strategy has met with complications in practice. First, there is no clear subset of putatively neutrally evolving introns; indeed, there is no consensus as to whether introns are generally beneficial, neutral, or deleterious or how the impact of introns on fitness might vary across lineages (Doolittle 1978; Lynch 2002). Second, the relevant molecular mutational mechanisms—in particular, those that lead to intron creation and loss from the genome—remain obscure and are known to be diverse (Llopart et al. 2002; Roy and Gilbert 2005; Stajich and Dietrich 2006; Irimia, Rukov, et al. 2008; Li et al. 2009; Roy and Irimia 2009a; Worden et al. 2009). Third, the genome-wide near absence of intron loss and gain over many millions of years in a variety of different groups of eukaryotes (e.g., vertebrates, and some genera of apicomplexans and fungi) suggests the possibility that intron loss and/or gain mutations simply do not occur in some lineages (Roy and Hartl 2006; Roy et al. 2006), in which case evolutionary conservation would not imply function.
The availability of many full genomes from diverse species allows us to circumvent these obstacles, at least in part. Here, we report the case of Meis homeobox genes. In contrast to the large amounts of intron loss and gain in some animal species observed in genome-wide comparisons (Seo et al. 2001; Rogozin et al. 2003; Edvardsen et al. 2004; Coulombe-Huntington and Majewski 2007b; Putnam et al. 2008), Meis genes have experienced almost no intron loss or gain in any studied species, particularly within the first ten exons of the gene. Unexpected conservation extends to the size of conserved introns: although intron size is thought to be relatively labile and not to persist over long evolutionary distances, Meis genes show clear conservation of relative intron sizes across genomically diverse species. Interestingly, long introns are known to show higher sequence conservation than short introns (Bergman and Kreitman 2001; Parsch 2003; Haddrill et al. 2005; Marais et al. 2005; Halligan and Keightley 2006; Parsch et al. 2010). The negative correlation between intron length and sequence divergence holds even within the set of longest introns, suggesting that the density of conserved sequence elements within introns may increase with intron length (Halligan and Keightley 2006). Thus, one possible explanation for the conserved long introns in Meis/hth is that they contain regulatory elements important for gene expression (Bergman and Kreitman 2001). Meis/hth genes are known to harbor the largest (or one of the largest) sets of associated highly conserved noncoding regions (HCNRs) in both vertebrates and in flies (Sandelin et al. 2004; Woolfe et al. 2005; Engstrom et al. 2007), with nearly a hundred associated HCNRs, depending on the study. Many of these HCNRs lie within these long introns (Engstrom et al. 2007; Visel et al. 2007; Dong et al. 2009; Xiong et al. 2009) and, in some cases, show even higher sequence conservation than the surrounding coding exons (Engstrom et al. 2007). Importantly, some of these elements have been shown to drive positive enhancer expression in reporter assays in mammals (Visel et al. 2007) or even to be involved in posttranscriptional regulation in dipterans (Glazov et al. 2005). Together, the conservation at the levels of intron loss/gain and intron size suggests that the introns in the 5′ region of Meis could encode conserved regulatory functions, leading to their retention across metazoans. Interestingly, the notion of Meis genes as hot spots for long-scale regulatory landscapes could extend beyond the transcribed regions: we also found that Meis genes are associated with large intergenic regions devoid of other genes (i.e., gene deserts) upstream and/or downstream Meis/hth genes in all studied metazoans (supplementary table S2), with the exceptions of some vertebrate Meis paralogs discussed above. As with intron lengths, longer intergenic regions are known to show less sequence divergence across species (Halligan and Keightley 2006).
Very Different Evolutionary Histories for 5′ and 3′ Regions of Meis Genes: Functional Causes and Consequences
In addition to general contrasts between Meis and genome-wide gene structure evolution, we found contrasting evolutionary histories for intron–exon structures of the 5′ and 3′ regions of Meis family genes (fig. 1A). As discussed above, the first 10 exons show remarkable conservation, with positions and relative sizes of the first 9 introns conserved across metazoans; by contrast, the 3′ of the gene shows a remarkable diversity of structures, evidencing intron and exon creation and loss, and great flexibility in AS patterns. These patterns echo findings in gene sequence evolution, in which different regions of the same protein may show opposed patterns of constraint or positive selection.
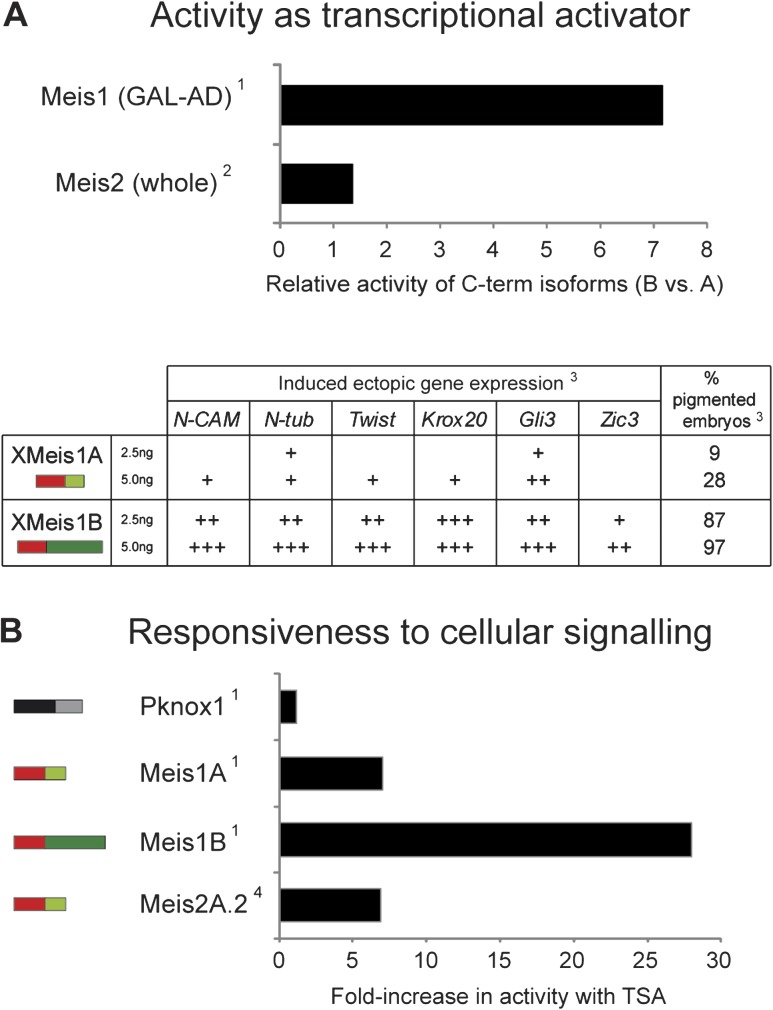
As with coding sequences, regional differences in protein function provide insight into the organismal functions undergoing potentially adaptive evolution. The conserved 5′ region encodes the highly conserved Pbx-interacting Meis (hth) domain and DNA-binding homeobox (Berthelsen et al. 1998; Mukherjee and Bürglin 2007), and the intervening introns may be implicated in developmental transcriptional regulation. By contrast, the variable 3′ region encodes interaction domains including the transcriptional activation domain and regulatory modules that modify protein transcriptional activity and response to cell signaling (Yang et al. 2000; Huang et al. 2005). For instance, regions within exons 11, 12a, and 12b affect transcriptional activator activity of human Meis1 proteins by mediating responsiveness to PKA and TSA (Huang et al. 2005; Shim et al. 2007), and inclusion of exon 12a lowers transcriptional activation in frogs and mammals (Yang et al. 2000; Maeda et al. 2001; Huang et al. 2005); (fig. 3). Shifting combinations of different isoforms and paralogs could thus allow subtle spatiotemporal control of Meis1 protein transcriptional activity (Huang et al. 2005; Heine et al. 2008; Sánchez-Guardado et al. 2011). Such modulation would be particularly powerful given MEIS proteins’ ability to enhance cell proliferation by transcriptional activation of cell cycle genes (Bessa et al. 2008; Heine et al. 2008): the quantitative combination of isoforms and paralogs present in each cell will likely affect the level of transcriptional activation of the target genes, and thus the proliferation rates. The 3′ region would therefore be a rich substrate for the evolution of different elaborations that could provide functional regulatory potential. Interestingly, this adaptation was likely aided by the gain of two introns in early bilaterians, splitting a single exon into three, allowing for a larger palette of potentially adaptive splicing-related mutations.
FIG. 3.
Summary of previous studies showing the different functional properties of C-terminal isoforms. (A) Differences in activity as transcriptional activators between C-terminal isoforms within each paralogous Meis gene in mammals. Top: histogram showing the ratio for activity of B (excluding 12a) versus A (including 12a) isoforms. Data for Meis1 correspond to a protein fusion of the activation domain (AD, C-terminus) from each of the isoforms. Meis2 data correspond to comparisons of full-length proteins and averaging the values for isoforms derived of inclusion/exclusion of exon 11′. Bottom: table summarizing the phenotypic results after injecting two different concentrations of full-length Meis1A or Meis1B isoforms in Xenopus embryos. Note that the intensity of the effect is not only isoform dependent but also concentration dependent. (B) Different C-terminal isoforms have different responses to TSA treatment. Histogram showing the fold-increase in transcriptional activation after TSA treatment for each Meis1 isoform, Meis2A.2 (excluding 11′ but including 12a) and the related pknox1 gene. References: (1) Huang et al. (2005), (2) Yang et al. (2000), (3) Maeda et al. (2001), and (4) Shim et al. (2007).
Deeply Conserved Splicing Functions in Bilaterians
Notably, our results show that Meis/hth genes harbor several cases of deeply conserved AS (Irimia et al. 2009). The AS of exon 10′ is a bilaterian innovation and has been maintained in some lineages since the very origin of bilaterians, representing one of the most ancestral AS events described to date (e.g., Mistry et al. 2003; Kalyna et al. 2006; Damianov and Black 2010). In addition, AS of exons 11 and 12a have likely arisen within chordate ancestors and have been conserved between amphioxus and vertebrates for some 600 My. This deep conservation of alternatively spliced sequences differs from the general low conservation of AS in different metazoan groups (reviewed in Irimia et al. 2009).
Frequent Gene Structural Convergence in Bilaterian Meis Evolution
Another striking result is the large number of cases of convergent evolution by identical or very similar sequence changes in different species in 3′ Meis regions. As with convergent protein changes in multiple lineages, these parallel changes suggest similar selective pressures acting in very different species; that these changes are restricted to the posttranscriptional regulatory domains of MEIS proteins suggests that evolution may have “used” and reused a finite set of accessible mechanisms for modulation of Meis function.
Recurrent Loss of Conserved Ancestral Sequence.
We find several cases of convergent loss of conserved ancestral sequences. First, the sequence of 3′ end of exon 10 (which we call 10′) is highly conserved across a wide variety of bilaterian and non-bilaterian genes (fig. 1C), indicating function; yet, this region has been independently lost from genomic copies of Meis genes at least nine times and is only very infrequently observed in transcripts of some genes that do contain it, begging the question of this sequence region's mode of function. Similarly, the coding sequence of exon 12b is conserved between studied non-bilaterians and various bilaterians but has been lost by constitutive protein truncations in three independent lineages (Pancrustacea, sea urchin and Meis3 in Anolis). In a third case, an ancestral vertebrate 5′ extension of exon 11 has been retained in many vertebrate genes but lost in 4 genic lineages; and the ancestral chordate AS exon 12a has been lost twice. Notably, these losses of conserved ancestral sequence have affected both putatively constitutively and alternatively spliced ancestral Meis gene regions.
Recurrent Evolution of Truncated C-Termini in Bilaterians.
One of the most striking observations concerns the regulation of the novel alternative exon 12a in vertebrates. Exon 12a is nearly constitutive in most vertebrate Meis genes, which is surprising because inclusion of this exon significantly attenuates transcriptional activation in vertebrate MEIS proteins, especially for Meis1 (Yang et al. 2000; Maeda et al. 2001; Huang et al. 2005). The scarce use of the most active, ancestral isoform may suggest that the emergence of exon 12a in chordates could have been associated with increasingly strict regulatory control of Meis target genes. This observation fits with postulates of the cybernetic theory of control in complex systems, which hold that the prevalence of negative regulatory mechanisms over positive ones increases system's stability (Wiener 1948). Notably, Ambulacraria and arthropods have independently achieved an equivalent situation by a different mechanism: evolution of constitutive 3′ extensions of exon 11 including premature stop codons. Insofar as these truncations also attenuate activator activity, these convergent patterns hint at an evolutionary trend towards more strictly regulated MEIS proteins in bilaterians.
The Fitness Effects of Introns and the Origins of Genome Complexity
Much discussion of spliceosomal introns has emphasized perspectives in which introns are neutral or slightly deleterious elements, potentially leading to evolutionary histories that are dominated by differences in mutation rates or effective population sizes (Lynch 2007). This study provides potential examples of two types of exceptions to this paradigm. First, widespread conservation of 5′ Meis intron–exon structures suggests strong purifying selection acting against intron loss in these regions. Second, the recurrent generation of very similar structures in the 3′ gene region suggests that alteration of intron–exon structures has been a frequent mechanism for adaptation. These findings join an increasingly long list of intronic loci encoding important organismal functions. Nonetheless, given the sheer number of introns in many metazoan genomes—reaching 200,000 in human and other vertebrate genomes—the proportion of introns whose loss is opposed by selection remains very much an open question, with answers ranging from a small minority to a clear majority still being well within the realm of possibility.
Concluding Remarks
The diversity of structures, functions, and mechanisms associated with transcript splicing, and uncertainty about the general fitness consequences of introns, complicate efforts to understand the function of individual introns. The current report details a case in which the evolution of one gene family contrasts strikingly with genome-wide patterns, suggesting purifying selection on intron–exon structures and suggesting functional roles for splicing in these genes. These results indicate the utility of many-species comparisons between introns within a genome. Future research should research toward explicit models for divergence in intron–exon structures among large sets of metazoans, to allow for systematic prediction of intron functionality across lineages and loci.
Supplementary Material
Supplementary figure S1–S6 and tables S1–S3 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Acknowledgments
We thank Salvatore D'Aniello for kindly helping with the experiments, José L. Gómez-Skarmeta for the X. tropicalis samples and Maria Ina Arnone for the S. purpuratus samples. M.I., I.M., and J.G.F. were funded by grants BFU2005-00252 and BMC2008-03776 from the Spanish Ministerio de Educación y Ciencia; M.I. and I.M. held FPI and FPU fellowships, respectively; M.H.S., by FUNDESALUD-PRIS09043 and BFU2010-19461; and L.P. and J.L.F. by BFU2008-04156 and SENECA Foundation contract 04548/GERM/06-10891.
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Alexander RP, Fang G, Rozowsky J, Snyder M, Gerstein MB. Annotating non-coding regions of the genome. Nat Rev Genet. 2010;11:559–571. doi: 10.1038/nrg2814. [DOI] [PubMed] [Google Scholar]
- Aparicio S, et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Azcoitia V, Aracil M, Martínez-A C, Torres M. The homeodomain protein Meis1 is essential for definitive hematopoiesis and vascular patterning in the mouse embryo. Dev Biol. 2005;280:307–320. doi: 10.1016/j.ydbio.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Bang ML, et al. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-ban linking system. Circ Res. 2001;89:1065–1072. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- Bergman CM, Kreitman M. Analysis of conserved noncoding DNA in Drosophila reveals similar constraints in intergenic and intronic sequences. Genome Res. 2001;11:1335–1345. doi: 10.1101/gr.178701. [DOI] [PubMed] [Google Scholar]
- Berthelsen J, Zappavigna V, Mavilio F, Blasi F. Prep1, a novel functional partner of Pbx proteins. EMBO J. 1998;17:1423–1433. doi: 10.1093/emboj/17.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessa J, et al. meis1 regulates cyclin D1 and c-myc expression, and controls the proliferation of the multipotent cells in the early developing zebrafish eye. Development. 2008;135:799–803. doi: 10.1242/dev.011932. [DOI] [PubMed] [Google Scholar]
- Choe SK, Vlachakis N, Sagerström CG. Meis family proteins are required for hindbrain development in the zebrafish. Development. 2002;129:585–595. doi: 10.1242/dev.129.3.585. [DOI] [PubMed] [Google Scholar]
- Caenorhabditis elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Sea Urchin Genome Sequencing Consortium et al. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribolium Genome Sequencing Consortium et al. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- Coulombe-Huntington J, Majewski J. Characterization of intron loss events in mammals. Genome Res. 2007a;17:23–32. doi: 10.1101/gr.5703406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe-Huntington J, Majewski J. Intron loss and gain in drosophila. Mol Biol Evol. 2007b;24:2842–2850. doi: 10.1093/molbev/msm235. [DOI] [PubMed] [Google Scholar]
- D'Aniello S, et al. Gene expansion and retention leads to a diverse tyrosine kinase superfamily in amphioxus. Mol Biol Evol. 2008;25:1841–1854. doi: 10.1093/molbev/msn132. [DOI] [PubMed] [Google Scholar]
- Damianov A, Black DL. Autoregulation of Fox protein expression to produce dominant negative splicing factors. RNA. 2010;16:405–416. doi: 10.1261/rna.1838210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Dibner C, Elias S, Frank D. XMeis3 protein activity is required for proper hindbrain patterning in Xenopus laevis embryos. Development. 2001;128:3415–3426. doi: 10.1242/dev.128.18.3415. [DOI] [PubMed] [Google Scholar]
- Dong X, Fredman D, Lenhard B. Synorth: exploring the evolution of synteny and long-range regulatory interactions in vertebrate genomes. Genome Biol. 2009;10:R86. doi: 10.1186/gb-2009-10-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF. Genes in pieces: were they ever together? Nature. 1978;272:581–582. [Google Scholar]
- Drummond A, Strimmer K. PAL: an object-oriented programming library for molecular evolution and phylogenetics. Bioinformatics. 2001;17:662–663. doi: 10.1093/bioinformatics/17.7.662. [DOI] [PubMed] [Google Scholar]
- Edvardsen RB, et al. Hypervariable and highly divergent intron/exon organizations in the chordate Oikopleura dioica. J Mol Evol. 2004;59:448–457. doi: 10.1007/s00239-004-2636-5. [DOI] [PubMed] [Google Scholar]
- Engstrom PG, Ho Sui SJ, Drivenes O, Becker TS, Lenhard B. Genomic regulatory blocks underlie extensive microsynteny conservation in insects. Genome Res. 2007;17:1898–1908. doi: 10.1101/gr.6669607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferran JL, Sánchez-Arrones L, Sandoval JE, Puelles L. A model of early molecular regionalization in the chicken embryonic pretectum. J Comp Neurol. 2007;505:379–403. doi: 10.1002/cne.21493. [DOI] [PubMed] [Google Scholar]
- Ghedin E, et al. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317:1756–1760. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazov EA, Pheasant M, McGraw EA, Bejerano G, Mattick JS. Ultraconserved elements in insect genomes: a highly conserved intronic sequence implicated in the control of homothorax mRNA splicing. Genome Res. 2005;15:800–808. doi: 10.1101/gr.3545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Haddrill PR, Charlesworth B, Halligan DL, Andolfatto P. Patterns of intron sequence evolution in Drosophila are dependent upon length and GC content. Genome Biol. 2005;6:R67. doi: 10.1186/gb-2005-6-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan DL, Keightley PD. Ubiquitous selective constraints in the Drosophila genome revealed by a genome-wide interspecies comparison. Genome Res. 2006;16:875–884. doi: 10.1101/gr.5022906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine P, Dohle E, Bumsted-O'Brien K, Engelkamp D, Schulte D. Evidence for an evolutionary conserved role of homothorax/Meis1/2 during vertebrate retina development. Development. 2008;135:805–811. doi: 10.1242/dev.012088. [DOI] [PubMed] [Google Scholar]
- Hellsten U, et al. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisa T, et al. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. 2004;23:450–459. doi: 10.1038/sj.emboj.7600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong X, Scofield DG, Lynch M. Intron size, abundance, and distribution within untranslated regions of genes. Mol Biol Evol. 2006;23:2392–2404. doi: 10.1093/molbev/msl111. [DOI] [PubMed] [Google Scholar]
- Huang H, et al. MEIS C termini harbor transcriptional activation domains that respond to cell signaling. J Biol Chem. 2005;280:10119–10127. doi: 10.1074/jbc.M413963200. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Hughes SS, Buckley CO, Neafsey DE. Complex selection on intron size in Cryptococcus neoformans. Mol Biol Evol. 2008;25:247–253. doi: 10.1093/molbev/msm220. [DOI] [PubMed] [Google Scholar]
- Hyman-Walsh C, Bjerke GA, Wotton D. An autoinhibitory effect of the homothorax domain of Meis2. FEBS J. 2010;277:2584–2597. doi: 10.1111/j.1742-464X.2010.07668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Maeso I, Garcia-Fernandez J. Convergent evolution of clustering of Iroquois homeobox genes across metazoans. Mol Biol Evol. 2008;25:1521–1525. doi: 10.1093/molbev/msn109. [DOI] [PubMed] [Google Scholar]
- Irimia M, Maeso I, Gunning PW, Garcia-Fernandez J, Roy SW. Internal and external paralogy in the evolution of Tropomyosin genes in metazoans. Mol Biol Evol. 2010;27:1504–1517. doi: 10.1093/molbev/msq018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Roy SW. Spliceosomal introns as tools for genomic and evolutionary analysis. Nucleic Acids Res. 2008;36:1703–1712. doi: 10.1093/nar/gkn012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, et al. Origin of introns by 'intronization' of exonic sequences. Trends Genet. 2008;24:378–381. doi: 10.1016/j.tig.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Irimia M, Rukov JL, Roy SW, Vinther J, Garcia-Fernandez J. Quantitative regulation of alternative splicing in evolution and development. Bioessays. 2009;31:40–50. doi: 10.1002/bies.080092. [DOI] [PubMed] [Google Scholar]
- Kalyna M, Lopato S, Voronin V, Barta A. Evolutionary conservation and regulation of particular alternative splicing events in plant SR proteins. Nucleic Acids Res. 2006;34:4395–4405. doi: 10.1093/nar/gkl570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Li W, Tucker AE, Sung W, Thomas WK, Lynch M. Extensive, recent intron gains in Daphnia populations. Science. 2009;326:1260–1262. doi: 10.1126/science.1179302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llopart A, Comeron JM, Brunet FG, Lachaise D, Long M. Intron presence-absence polymorphism in Drosophila driven by positive Darwinian selection. Proc Natl Acad Sci U S A. 2002;99:8121–8126. doi: 10.1073/pnas.122570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon J. Worm genomes hold the smoking guns of intron gain. Proc Natl Acad Sci U S A. 2004;101:11195–11196. doi: 10.1073/pnas.0404148101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. The Origins of Genome Architecture. Sunderland (MA): Sinauer Associates; 2007. [Google Scholar]
- Lynch M. Intron evolution as a population-genetic process. Proc Natl Acad Sci U S A. 2002;99:6118–6123. doi: 10.1073/pnas.092595699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda R, et al. Xmeis1, a protooncogene involved in specifying neural crest cell fate in Xenopus embryos. Oncogene. 2001;20:1329–1342. doi: 10.1038/sj.onc.1204250. [DOI] [PubMed] [Google Scholar]
- Marais G, Nouvellet P, Keightley PD, Charlesworth B. Intron size and exon evolution in Drosophila. Genetics. 2005;170:481–485. doi: 10.1534/genetics.104.037333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercader N, et al. Conserved regulation of proximodistal limb axis development by Meis1/Hth. Nature. 1999;402:425–429. doi: 10.1038/46580. [DOI] [PubMed] [Google Scholar]
- Mercader N, Tanaka EM, Torres M. Proximodistal identity during vertebrate limb regeneration is regulated by Meis homeodomain proteins. Development. 2005;132:4131–4142. doi: 10.1242/dev.01976. [DOI] [PubMed] [Google Scholar]
- Mistry N, Harrington W, Lasda E, Wagner EJ, Garcia-Blanco MA. Of urchins and men: evolution of an alternative splicing unit in fibroblast growth factor receptor genes. RNA. 2003;9:209–217. doi: 10.1261/rna.2470903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe D. Genetics. Genomic clues to DNA treasure sometimes lead nowhere. Science. 2009;325:142–143. doi: 10.1126/science.325_142. [DOI] [PubMed] [Google Scholar]
- Moskow JJ, Bullrich F, Huebner K, Daar IO, Buchberg AM. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol Cell Biol. 1995;15:5434. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K, Bürglin TR. Comprehensive analysis of animal TALE homeobox genes: new conserved motifs and cases of accelerated evolution. J Mol Evol. 2007;65:137–153. doi: 10.1007/s00239-006-0023-0. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Jenkins NA, Copeland NG. Identification of a new family of Pbx-related homeobox genes. Oncogene. 1996;13:2235–2242. [PubMed] [Google Scholar]
- Ng L, et al. An anatomic gene expression atlas of the adult mouse brain. Nat Neurosci. 2009;12:356–362. doi: 10.1038/nn.2281. [DOI] [PubMed] [Google Scholar]
- Noro B, Culi J, McKay DJ, Zhang W, Mann RS. Distinct functions of homeodomain-containing and homeodomain-less isoforms encoded by homothorax. Genes Dev. 2006;20:1636–1650. doi: 10.1101/gad.1412606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oulad-Abdelghani M, et al. Meis2, a novel mouse Pbx-related homeobox gene induced by retinoic acid during differentiation of P19 embryonal carcinoma cells. Dev Dyn. 1997;210:173–183. doi: 10.1002/(SICI)1097-0177(199710)210:2<173::AID-AJA9>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Pai CY, et al. The homothorax homeoprotein activates the nuclear localization of another homeoprotein, extradenticle, and suppresses eye development in Drosophila. Genes Dev. 1998;12:435–446. doi: 10.1101/gad.12.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch J. Selective constraints on intron evolution in Drosophil. Genetics. 2003;165:1843–1851. doi: 10.1093/genetics/165.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch J, Novozhilov S, Saminadin-Peter SS, Wong KM, Andolfatto P. On the utility of short intron sequences as a reference for the detection of positive and negative selection in Drosophila. Mol Biol Evol. 2010;27:1226–1234. doi: 10.1093/molbev/msq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann M, Prpic NM. Appendage patterning in the South American bird spider Acanthoscurria geniculata (Araneae: mygalomorphae) Dev Genes Evol. 2009;219:189–198. doi: 10.1007/s00427-009-0279-7. [DOI] [PubMed] [Google Scholar]
- Prpic NM, Janssen R, Wigand B, Klingler M, Damen WG. Gene expression in spider appendages reveals reversal of exd/hth spatial specificity, altered leg gap gene dynamics, and suggests divergent distal morphogen signaling. Dev Biol. 2003;264:119–140. doi: 10.1016/j.ydbio.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Putnam N, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Putnam NH, et al. Sea anemone genome reveals ancestral Eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Rieckhof GE, Casares F, Ryoo HD, Abu-Shaar M, Mann RS. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Wolf YI, Sorokin AV, Mirkin BG, Koonin EV. Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr Biol. 2003;13:1512–1517. doi: 10.1016/s0960-9822(03)00558-x. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Roy SW, Fedorov A, Gilbert W. Large-scale comparison of intron positions in mammalian genes shows intron loss but no gain. Proc Natl Acad Sci U S A. 2003;100:7158–7162. doi: 10.1073/pnas.1232297100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Gilbert W. The pattern of intron loss. Proc Natl Acad Sci U S A. 2005;102:713–718. doi: 10.1073/pnas.0408274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Hartl DL. Very little intron loss/gain in Plasmodium: intron loss/gain mutation rates and intron number. Genome Res. 2006;16:750–756. doi: 10.1101/gr.4845406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Irimia M. Mystery of intron gain: new data and new models. Trends Genet. 2009a;25:67–73. doi: 10.1016/j.tig.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Roy SW, Irimia M. Splicing in the eukaryotic ancestor: form, function and dysfunction. Trends Ecol Evol. 2009b;24:447–455. doi: 10.1016/j.tree.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Roy SW, Irimia M, Penny D. Very little intron gain in Entamoeba histolytica genes laterally transferred from prokaryotes. Mol Biol Evol. 2006;23:1824–1827. doi: 10.1093/molbev/msl061. [DOI] [PubMed] [Google Scholar]
- Roy SW, Penny D. Large-scale intron conservation and order-of-magnitude variation in intron loss/gain rates in apicomplexan evolution. Genome Res. 2006;16:1270–1275. doi: 10.1101/gr.5410606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rukov JL, et al. High qualitative and quantitative conservation of alternative splicing in Caenorhabditis elegans and Caenorhabditis briggsae. Mol Biol Evol. 2007;24:909–917. doi: 10.1093/molbev/msm023. [DOI] [PubMed] [Google Scholar]
- Sánchez-Guardado LÓ, et al. Distinct and redundant expression and transcriptional diversity of Meis gene paralogs during chicken development. Dev Dyn. 2011;240:1475–1492. doi: 10.1002/dvdy.22621. [DOI] [PubMed] [Google Scholar]
- Sandelin A, et al. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics. 2004;5:99. doi: 10.1186/1471-2164-5-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker D, et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- Scofield DG, Hong X, Lynch M. Position of the final intron in full-length transcripts: determined by NMD? Mol Biol Evol. 2007;24:896–899. doi: 10.1093/molbev/msm010. [DOI] [PubMed] [Google Scholar]
- Seo H-C, et al. Miniature genome in the marine chordate Oikopleura dioica. Science. 2001;294:2506. doi: 10.1126/science.294.5551.2506. [DOI] [PubMed] [Google Scholar]
- Shim S, Kim Y, Shin J, Kim J, Park S. Regulation of EphA8 gene expression by TALE homeobox transcription factors during development of the mesencephalon. Mol Cell Biol. 2007;27:1614–1630. doi: 10.1128/MCB.01429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- Stajich JE, Dietrich FS. Evidence of mRNA-mediated intron loss in the human-pathogenic fungus Cryptococcus neoformans. Eukaryot Cell. 2006;5:789–793. doi: 10.1128/EC.5.5.789-793.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toresson H, Parmar M, Campbell K. Expression of Meis and Pbx genes and their protein products in the developing telencephalon: implications for regional differentiation. Mech Dev. 2000;94:183–187. doi: 10.1016/s0925-4773(00)00324-5. [DOI] [PubMed] [Google Scholar]
- Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz AJ, Rikhof HA, Hernandez RE, Moens CB. Zebrafish Meis functions to stabilize Pbx proteins and regulate hindbrain patterning. Development. 2001;128:4139–4151. doi: 10.1242/dev.128.21.4139. [DOI] [PubMed] [Google Scholar]
- Wassef MA, et al. Rostral hindbrain patterning involves the direct activation of a Krox20 transcriptional enhancer by Hox/Pbx and Meis factors. Development. 2008;135:3369–3378. doi: 10.1242/dev.023614. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston R, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wiener N. Cybernetics or control and communication in the animal and the machine. New York: John Wiley & Sons Inc; 1948. [Google Scholar]
- Williams TM, Williams ME, Innis JW. Range of HOX/TALE superclass associations and protein domain requirements for HOXA13:MEIS interaction. Dev Biol. 2005;277:457–471. doi: 10.1016/j.ydbio.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Woolfe A, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden AZ, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- Xiong L, et al. MEIS1 intronic risk haplotype associated with restless legs syndrome affects its mRNA and protein expression levels. Hum Mol Genet. 2009;18:1065–1074. doi: 10.1093/hmg/ddn443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, et al. Three-amino acid extension loop homeodomain proteins Meis2 and TGIF differentially regulate transcription. J Biol Chem. 2000;275:20734–20741. doi: 10.1074/jbc.M908382199. [DOI] [PubMed] [Google Scholar]