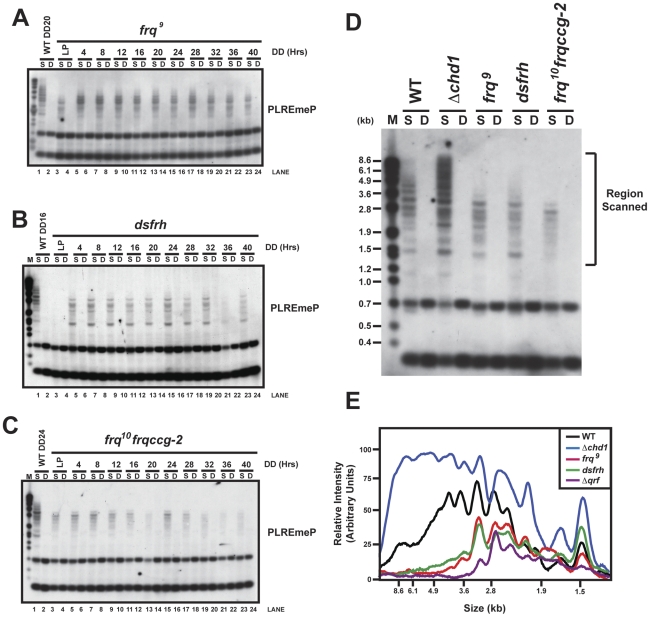
Figure 6. FRQ and the antisense transcript are required for normal DNA methylation.
(A)The methylation status of the frq promoter was examined in frq9, (B) a FRH knock-down strain dsfrh, and (C) a strain with reduced qrf expression, frq10frqccg-2. Strains were grown in light, transferred to darkness, samples collected at the indicated times, DNA was isolated and digested with Sau3AI (S) or DpnII (D) and processed for Southern hybridizations using a PLRE specific probe. WT DNA isolated from dark grown cultures was included as a control to compare differences in migration of methylated DNA. (D) To obtain a side-by-side comparison of the methylation pattern for the mutant strains used in this study, DNA obtained from the DD16 time point was taken for each strain and treated as in panel A, B and C. (E) The level of DNA methylation in the samples from (D) was quantified by performing densitometry on the region indicated in (D) and plotted versus the molecular weight marker. It is clear from this figure that Δchd1 is hypermethylated whereas frq9, dsfrh, and frq10frqccg-2 are all hypomethylated relative to WT.