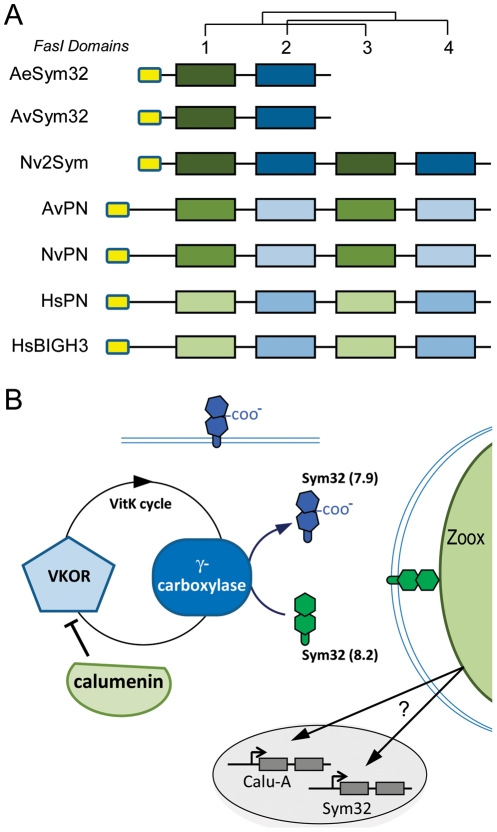
Figure 7. Sym32 gene duplication and putative γ-carboxylation regulation model.
A. Color code schematization of the PhyML tree for the FasI domains of the Sym32, Periostin (PN) and BGH3 homologs in sea anemones (Ae, A. elegantissima; Av, A. viridis; Nv, N. vectensis) and human (Hs) (see tree in Figure S7b). Each rectangle represents one FasI domain (small yellow rectangles represent the Signal Sequence). FasI domains 1 and 3 are closely related. The first FasI domains of [AeSym32-1, AvSym32-1 and Nv2Sym-1 and -3] are closely related to each other and to a lesser extent with [AvPN-1 and -3, and NvPN-1 and -3] and finally with [HsPN-1 and -3, and HsBGH3-1 and -3]. A similar relationship exists for the FasI domains 2 and 4. AeSym32, AvSym32 and Nv2Sym are probable orthologs, except that Sym32 appears as the first half of Nv2Sym. The putative ortholog of HsPN is NvPN. B. Heuristic Model. The presence of symbionts activates the expression of calumenin and sym32 genes via an unknown mechanism. The CRS of the Sym32 protein is recognized as substrate by the activated vitamin K cycle (vitamin K is a cofactor produced from the photosynthetic organism) and in turn is γ-carboxylated. Meanwhile, the Calumenin represses the VKOR protein, inhibiting the γ-carboxylase. Two forms of Sym32 are thus expected to be produced from this pathway: the Glu-Sym32 and Gla-Sym32 electrophoretypes (likely corresponding to the two spots PI = 8.2 and PI = 7.9, respectively [51]). Only Sym32 (PI = 8.2) is found to be associated with the symbiosome membrane, underlying a novel functionality for Sym32 and γ-carboxylation.