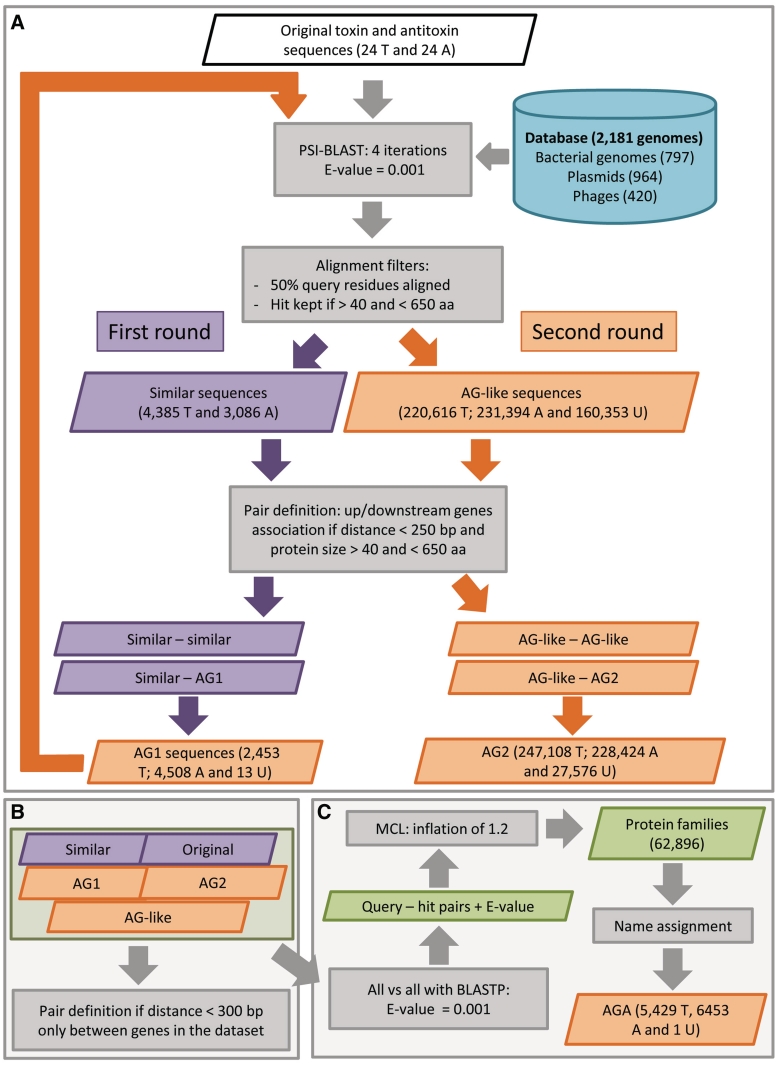
Figure 1.
Association by guilt bioinformatics approach. Detection of ‘associated by guilt’ (AG) sequences (A). ‘Similar’ sequences (purple box) to the 48 ‘original’ sequences (white box) were detected by PSI-BLAST searches using the indicated parameters (gray boxes) and a database composed of 2181 genome sequences (blue cylinder). First round: AG toxins and antitoxins denoted as AG1 (orange box) were identified by paring of the ‘similar’ sequences using the indicated parameters (gray box). Number and type of sequences are indicated (T for toxin, A for antitoxin and U for unassigned). Second round: AG1 sequences were used as query for a second round of detection using the same parameters as in the initial steps to identify AG-like sequences (orange box). AG2 sequences were identified by pairing the AG-like sequences. Number and type of sequences are indicated. Pair definition (B). An additional round of pair definition was performed using sequences detected in (A) using the indicated parameters (gray box). Protein families (C). Protein families (green box) were generated by grouping all the proteins present in the dataset using the Markov clustering algorithm (MCL) with the indicated parameters (gray boxes). Names of ‘original’ or ‘similar’ sequences were propagated to the AG proteins within the same protein family. These sequences were then defined as ‘associated by guilt and annotated’ (AGA).