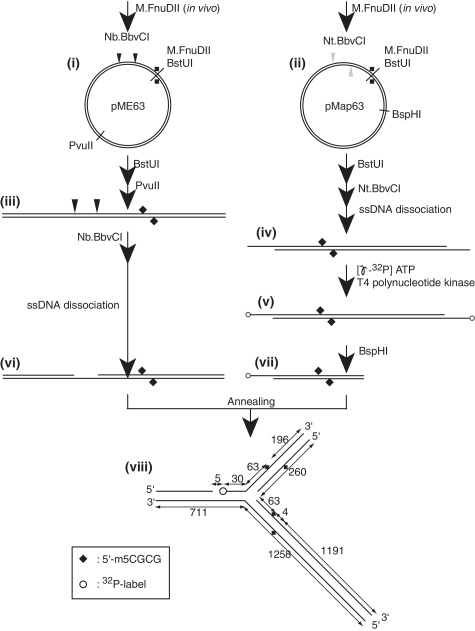
Figure 2.
Preparation of a long-branched DNA with methylation. The two plasmids were modified in vivo by M.FnuDII to generate 5′-m5CGCG, a McrBC recognition sequence {(i), (ii)}. Potential unmethylated plasmids were eliminated by cleavage with BstUI (5′-CGCG). pME63 was cleaved with PvuII and then with nicking endonuclease Nb.BbvCI (iii), while pMap63 was treated with nicking endonuclease Nt.BbvCI (iv). The resulting short single strands were dissociated by heating and removed by annealing with a complementary single-strand oligo DNA. The 5′-ends of intermediate (iv) were labeled with 32P (v), followed by BspHI cleavage for removal of one of the radio-labels (vii). The two DNAs with complementary single-strand regions {(vi), (vii)} were annealed to form a branched structure {viii, eM63(++)} as detailed in ‘Materials and Methods’ section. Open circle, 32P label at 5′-end; filled diamond, DNA methylation.