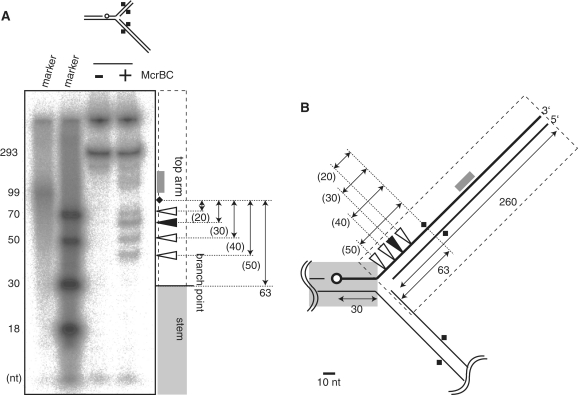
Figure 4.
Mapping cleavage sites. (A) A model replication fork (eM63++) cleaved by McrBC was subjected to polyacrylamide gel electrophoresis under denaturatingconditions (6M urea). (B) Cleavage sites. Open circle, 32P-label at 5′-end; Square, DNA methylation; Filled triangle, major product; white triangle, minor product; gray box, products corresponding to the smear. There are two labeled bands in the untreated lane, corresponding to the strand participating in the fork (293 nt), and not denaturated materials stacked in the well.