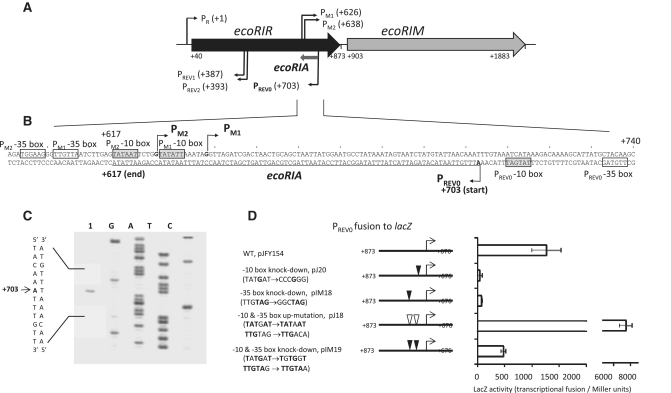
Figure 1.
The antisense RNA (Rna0) encoded by the ecoRIA gene. (A) EcoRI restriction–modification system. Coordinates are relative to the transcription initiation site for the bicistronic ecoRIRM mRNA from PR promoter. PM1 and PM2, two overlapping promoters for ecoRIM transcription (28); PREV1 and PREV2, two overlapping reverse promoters associated with negative effects on expression from the PR promoter (29); PREV0, a reverse promoter for an antisense RNA studied in this report. (B) ecoRIA antisense RNA gene. Transcription initiation sites are indicated by arrows. The ecoRIA sequence is underlined. Promoter hexamers are boxed in gray (−10 box) and white (−35 box) (11, this work). (C) Mapping the transcription initiation site by primer extension. Extension from a lacZ gene-specific primer (lacP) (Supplementary Table S1) was carried out using total RNA from E. coli harboring pJFY154 (lane 1, see below). Lanes G, A, T and C represent products of the dideoxy sequencing reactions carried out with the same primer. The initiation site is indicated by an arrow. (D) Promoter activity of PREV0. DNA fragment carrying PREV0 was fused to the lacZ gene (pJFY154). Substitutions were made at the −35 (pIM18) or −10 box (pJ20) of the PREV0 promoter to knock down promoter activity, as well as to increase its activity, by matching them to the consensus sequences for E. coli σ70 RNA polymerase (pJ18). PREV0 activity with the 3 nt changes present in pIM24 (Table 1) was also measured (pIM19) (see Figure 4 below). These changes did not affect the amino acid sequence. Each value represents an average of four measurements, along with their standard deviations.