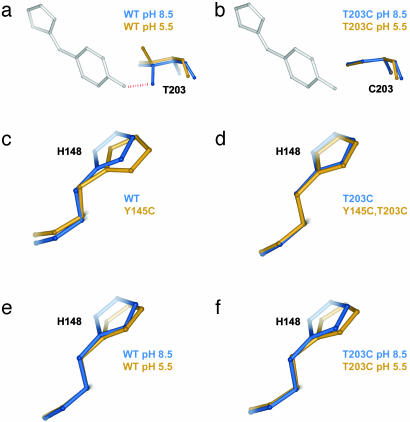
Fig. 5.
Specific examples of the structural cycle analysis. (a and b) The effect of ΔpH at position 203 in the wild-type (a) or T203C (b) mutant background and illustrates a mechanistically clear case of structural nonadditivity (coupling). T203 is hydrogen-bonded to the phenolic oxygen of the chromophore at pH 8.5, and protonation of this site upon pH shift breaks the bond and causes T203 to rotate away by 120°. However, cysteine at 203 is not hydrogen bonded at either pH, and consequently shows no structural change upon ΔpH. Thus, the structural coupling at position 203 reflects the fact that either ΔpH or T203C eliminates the hydrogen bond, and the double mutant has no further effect than that of each mutant taken independently. (c and d) A less predictable example of structural coupling in the Y145C–T203C cycle. H148 is displaced by Y145C in the wild-type background (c) but is not displaced in the T203C background (d); this T203C dependence in the repacking of H148 induced by Y145C is the basis of the observed structural coupling between these two mutations. (e and f) An example of structural additivity in the T203C–ΔpH cycle. H148 is displaced by ΔpH (e), but this displacement is the same in the background of T203C (f); thus, T203C and ΔpH fail to show structural coupling at this site. This result is particularly striking because T203C itself displaces H148 (Fig. 2a).