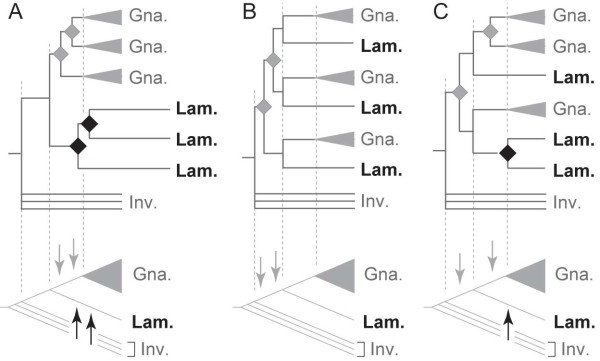
Figure 1.
Alternative tree toplogies supporting different timings of two-round whole genome duplications (2R-WGDs). As an example, for a gene family with three gnathostome paralogs and three lamprey paralogs, gene trees (top) and species trees with timings of gene duplications (bottom) are shown. Gene duplications that gave rise to multiple gnathostome paralogs are indicated with grey diamond (top) and grey arrows (bottom), and those in the lamprey lineage are indicated with black diamond (top) and black arrows (bottom). Even though a recent large-scale phylogenetic analysis supported the scenario in B [4], analyses on single gene families often result in the tree topology similar to that in A. C was previously supported [49]. Abbreviations: Gna., gnathostome gene; Lam., lamprey gene; Inv., invertebrate gene.