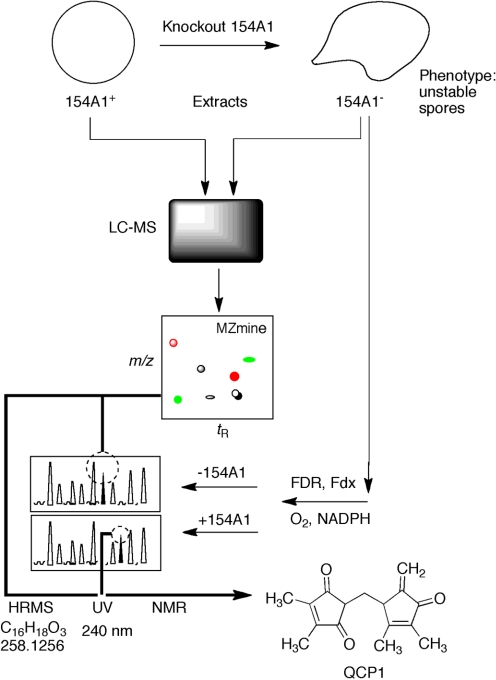
Fig. 4.
Use of an MZmine approach to identification of a substrate and product for S. coelicolor P450 154A1. LC-MS data were obtained for wild-type and P450 154AA1-knockout bacteria. Comparison of the LC-MS data by LC-MS using MZmine suggested a compound with MH+ 259. Extracts of the P450 154A1 bacteria (in which a substrate would be expected to accumulate) with P450 154A1 indicated that the candidate compound disappeared during incubation and a new peak appeared. Subsequent analytical work showed 1) the structures of the substrate QCP1 and product QCP2a,b (2 isomers) and 2) that no redox function was related to the change (Fig. 5) (Cheng et al., 2010).