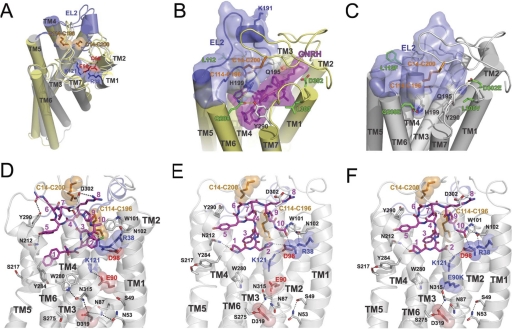
Fig. 1.
A, two Cys bridges (Cys14-Cys200 and Cys114-Cys196) and two ionic pairs (Arg38-Asp98 and Glu90-Lys121) in homology model of human GnRH receptor in the inactive conformation. Inactive (gray) and active (yellow) states were modeled using rhodopsin crystal structures (Protein Data Bank codes 1U19 and 3DQB, respectively). EL2 loops (residues 181–204) are colored blue, Cys residues are colored orange, Arg and Lys residues are colored blue, and Asp and Glu residues are colored red. Superposition of active and inactive conformation shows that the largest conformational changes include outward rigid body movement of TM6, movement of TM5 toward TM6, tilting of TM7, and shift of its middle part toward TM2. B and C, the proposed movement of the EL2 in the wild-type and mutant human GnRH receptor. B, homology models of the wild-type human receptor (yellow) in active state with GnRH decapeptide (purple) show the upper position of the EL2 (blue), which is attached to the receptor by two disulfide bridges (Cys14-Cys200 and Cys114-Cys196). C, homology model of the ΔLys191, L112F, Q208E, L300V, and D302E mutant of the human receptor in the inactive state (gray) with EL2 loop (blue) inserted between helices and filling the ligand binding pocket. Deletion of charged Lys191 permits the movement of EL2 inside the seven-helical bundle. Substitutions in noncontiguous four-residue motif that interacts with EL2 (Gln208 forms H-bond with His199, Val300 allows reorientation of Tyr290 that interacts with His199, and Glu302 forms H-bond with Gln195) may facilitate the inward movement of EL2. Multiple interaction of EL2 with helices may stabilize the receptor structure. D to F, rearrangement of ionic bridges and H-bond networks during activation of wild-type and mutant human GnRH receptor. D, model of the inactive conformation of receptor in complex with peptide antagonist cetrorelix. Two ionic bridges are formed between TM1-TM2 (Arg38-Asp98) and TM2-TM3 (Glu90-Lys121). The H-bond network that is formed in TM1-TM3-TM6-TM7 (Asn53-Asn87-Asp319-Asp315-Trp280) additionally stabilizes the receptor structure. Antagonist interacts with Arg38-Asp98 salt bridge, but not with Lys121. E, model of the active conformation of the receptor complexed with the natural decapeptide agonist GnRH. The ionic bridge Glu90-Lys121 is broken, and Lys121 together with Asp98, interact with His2 of GnRH. Arg38 forms H-bond with Gly10-NH2 of the ligand. Rearranged residues (Asn53-Ser49 and Asn87-Glu90-Asn315-Glu319-Ser275) form H-bond network in TM1-TM2-TM3-TM6-TM7. Tyr284 rotates and forms H-bond with Ser217. F, model of the E90K mutant of human GnRH receptor in complex with Asp2-GnRH decapeptide, which acts as an agonist. Salt bridge Glu90-Lys121 is broken, and Lys121 interacts with Asp2 of the ligand and also forms an ionic pair with Asp98. H-bond network is similar to one in the active conformation of the wild-type receptor. Lys90 is involved not only in H-bonding with Asn315 but also in the ionic attraction with Asp319 and repulsion with Lys121. These ionic and H-bonding interactions of Lys90 probably stabilize the active conformation, which leads to CA. Interaction of decapeptide ligands (colored purple) in all three models are in good agreement with the experimental data.