Fig. 1.
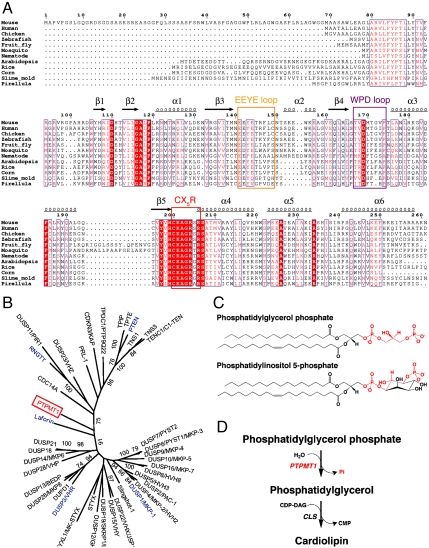
PTPMT1 functions in the cardiolipin biosynthesis pathway. (A) Structure-based sequence alignment of PTPMT1. The EEYE loop, WPD loop, and CX5R residues are highlighted with orange, magenta, and red rectangles, respectively. Highly conserved residues are highlighted in red. Secondary structural elements are shown above the sequence block. Pirellula is a marine bacterium. (B) A phylogenetic tree of human DSPs built using maximum likelihood (ML) method in MEGA5.0. The tree was calculated based on the DSP domain regions only, using Poisson Correction model and partial deletion of gaps. Each branch was tested by 100 bootstrap replicates, and the branches with bootstrap values above 70% were shown. Proteins mentioned in the paper, including PTPMT1, Laforin, RNGTT, PTEN, VHR, and MKP-1 are highlighted in red and blue. (C) Chemical structures of PGP and PI(5)P. The head group of PGP and equivalent regions in PI(5)P are highlighted in red. (D) PTPMT1 dephosphorylates PGP in the cardiolipin biosynthesis pathway (shown in red).