Abstract
OBJECTIVE
Macrophage secretion of proinflammatory cytokines contributes to the pathogenesis of obesity-related insulin resistance. An important regulator of inflammation is the suppressor of cytokine signaling-1 (SOCS1), which inhibits the JAK-STAT and toll-like receptor-4 (TLR4) pathways. Despite the reported role of SOCS1 in inhibiting insulin signaling, it is surprising that a SOCS1 polymorphism that increases SOCS1 promoter activity is associated with enhanced insulin sensitivity despite obesity. In the current study, we investigated the physiological role of myeloid and lymphoid cell SOCS1 in regulating inflammation and insulin sensitivity.
RESEARCH DESIGN AND METHODS
We used mice generated by crossing SOCS1 floxed mice with mice expressing Cre recombinase under the control of the LysM-Cre promoter (SOCS1 LysM-Cre). These mice have deletion of SOCS1 in macrophages and lymphocytes. We assessed macrophage inflammation using flow cytometry and serum cytokine levels using Bioplex assays. We then measured insulin sensitivity using glucose tolerance tests and the euglycemic-hyperinsulinemic clamp. Using bone marrow–derived macrophages, we tested the effects of SOCS1 deletion in regulating responses to the TLR4 ligands: lipopolysaccharide (LPS) and palmitic acid.
RESULTS
SOCS1 LysM-Cre mice had increased macrophage expression of CD11c, enhanced sensitivity to LPS, and palmitic acid and increased serum concentrations of tumor necrosis factor-α, interleukin-6, and monocyte chemoattractant protein. Increased inflammation was associated with impaired glucose tolerance and hyperinsulinemia as a result of reduced hepatic but not skeletal muscle insulin sensitivity.
CONCLUSIONS
The expression of SOCS1 in hematopoietic cells protects mice against systemic inflammation and hepatic insulin resistance potentially by inhibiting LPS and palmitate-induced TLR4 signaling in macrophages.
The link between inflammation and diabetes was discovered more than a century ago when high doses of sodium salicylate were shown to ameliorate glycosuria (as reviewed in 1). Today, it is well established that obesity is associated with increased levels of proinflammatory cytokines that lead to insulin resistance (2) and defects in fatty acid metabolism (3). Accumulation of inflamed macrophages within adipose tissue is the major source of cytokines in obesity (4,5), and their activation has been linked to the Toll-like receptor (TLR) signaling family, which are known to activate proinflammatory signaling pathways. The best characterized member of this family, TLR4, binds to lipopolysaccharide (LPS) of Gram-negative bacterial cell walls and recruits the myeloid differentiation factor 88 to the Toll/interleukin (IL)-1 receptor domain of the receptor. This triggers the activation of the nuclear factor (NF)-κB pathway resulting in the expression of proinflammatory cytokines such as tumor necrosis factor (TNF)-α, IL-6, and IL-1β (6,7). In obesity, alterations in gut microbial flora that increase the endotoxin (LPS) load in the intestine have been postulated as a potential mechanism leading to obesity-related inflammation, insulin resistance, and nonalcoholic fatty liver disease (8,9). Alternatively, abrogation of LPS signaling in CD14 (a coreceptor of TLR4) mutant mice or reducing the content of gram-negative bacteria reduces inflammation and improves insulin resistance (10).
In addition to classical activation of the TLR signaling pathway via LPS several studies have demonstrated that saturated fatty acids such as palmitic acid also induce an inflammatory response in macrophages in part through activation of TLR signaling (11,12). Importantly, in mice deficient for both TLR2 and TLR4 obesity and free fatty acid–induced inflammation and insulin resistance is markedly blunted (11,13). These studies provide a compelling molecular connection for the strong correlation between inflammatory cytokines and circulating free fatty acids in obesity (14) and support findings demonstrating that the acute infusion of saturated fatty acids dramatically increases circulating TNF-α levels (15,16).
The suppressor of cytokine signaling (SOCS) family of proteins (SOCS1-SOCS7 and cytokine–inducible SH-2-containing protein) are induced by proinflammatory cytokines and negatively regulate cytokine signaling (17). SOCS1 inhibits signaling by interferons (18) and by cytokines that signal through the common-γ chain receptor, such as IL-2 (19) and IL-4 (20). SOCS1 has also been shown to negatively regulate TLR4 signaling by mediating the degradation of the adaptor protein Mal (21), which is involved in the transactivation of NF-κB after TLR4 ligation. In support of these findings genetic disruption of SOCS1 results in the dramatic dysregulation of immune and inflammatory responses (22,23).
Genetic polymorphisms in the promoter regions of the SOCS1 gene that influence its transcriptional activity have been described in humans (24,25) and the rs33977706 (−820 G>T) and rs243330 (−1456 G>A) polymorphisms are associated with improved insulin sensitivity. It is noteworthy that the rs33977706 (−820 G>T) polymorphism in the SOCS1 promoter is also associated with increased transcriptional activity. Increased SOCS1 expression associated with improved insulin sensitivity would at first seem to be at odds with the reported role of SOCS1 by us and others as a negative regulator of hepatic insulin sensitivity, effects that are mediated through downregulation and ubiquination of the insulin receptor substrate (IRS)-2 (26,27). However, we have shown that the deletion of SOCS1 from myeloid and lymphoid cells in SOCS1 LysM-Cre mice increases inflammation (23), and we hypothesize that this may lead to insulin resistance. In the current study, we used these mice and found they develop hyperinsulinemia and hepatic insulin resistance. Furthermore, deletion of SOCS1 in macrophages increased their sensitivity to LPS and palmitic acid. Therefore, we speculate that the hypersensitivity of SOCS1-deficient macrophages to LPS and palmitic acid may be a contributing factor to the increased inflammation and dysregulation in liver insulin sensitivity observed in SOCS1 LysM-Cre mice.
RESEARCH DESIGN AND METHODS
Animal experiments.
The animal ethics committee of St. Vincent's Health approved all procedures. SOCS1 LysM-Cre mice and their littermate controls (SOCS1fl/fl) have been described previously (23). All studies were performed in male mice aged 12–15 weeks of age. SOCS1fl/− and SOCS1fl/fl mice were generated on a C57BL/6 background, and LysM-Cre mice were on their 10th generation backcross to C57BL/6 before crossing to SOCS1fl/fl mice. Mice were maintained with ad libitum access to food and water in micro isolators cages, under pathogen free conditions with a 12-h light/dark cycle. Blood glucose, insulin, and cytokines were measured after a 6-h fast or in randomly fed animals. Glucose tolerance tests (GTT) were performed in conscious mice after a 6-h fast. For GTTs, mice were injected with 1.0 g/kg of D-glucose (Sigma) in 0.9% saline i.p. and blood glucose in tail vein samples was measured at 0, 15, 30, 45, 60, 90, and 120 min using Accuchek Advantage II Glucose Strips and Advantage glucometer (Roche Diagnostics, Mannheim, Germany) as described (28).
Euglycemic-hyperinsulinemic clamp.
Clamps were performed in conscious mice fasted for 6 h as recently described (28,29). Clamp insulin was measured by ELISA (Mercodia, Diagenics, U.K.).
Analytical methods
Bone marrow–derived macrophages.
Bone marrow–derived macrophages from SOCS1 LysM-Cre and control littermates were prepared as follows. In brief, mononuclear phagocyte progenitor cells derived from femoral and tibial bone marrow were cultured and propagated at 1 × 106 cells/mL (20 mL) in tissue-culture flasks with macrophage maintenance media consisting of L929-cell conditioned medium (containing M-CSF), 10% fetal calf serum, and RPMI 1640 medium. After day 1, cells were transferred to sterile 10-cm.
Petri dishes (10 mL/plate) and split and passaged into more Petri dishes between days 3 and 5 once they became confluent. On day 6, nonadherent cells were washed away with sterile PBS, and adherent cells were liberated with warm, Versene PBS (EDTA/PBS) and replated out at 2 × 106 cells/mL in 6-well tissue-culture plates. Cells were starved of serum overnight and treated with LPS (at 0, 1, 10, 100, and 1,000 ng/mL; Sigma, St. Louis, MO) or palmitate (0, 10, 100, 250, 500, and 1,000 μmol/L; Sigma) for 4 h. The cells were then lysed in TRIzol (Invitrogen, Carlsbad, CA), and RNA was extracted for quantitative RT-PCR.
Muscle 2-deoxy-D-glucose uptake.
Glucose uptake experiments in isolated extensor digitorum longus muscles were completed as recently described (30). In brief, muscles were treated with or without 2.8 μmol/L insulin (Actrapid; Novo Nordisk, Bagsværd, Denmark), and radioactivity was measured by liquid scintillation counting (Tri-Carb 2000; Packard Instruments, Meriden, CT).
Isolation and preparation of peritoneal macrophages for flow cytometry.
Resident peritoneal cells were harvested from control and SOCS1 LysM-Cre mice by flushing the peritoneal cavity with 10 mL PBS and immediately centrifuged at 500 g for 5 min at 4°C. For cell sorting, the pellet was resuspended in 1 mL red blood cell lysis buffer (eBioscience, San Diego, CA) and incubated for 5 min before resuspension in PBS containing 0.5% endotoxin-free BSA at a concentration of 107 cells/mL. Cells were incubated with fluorophore-conjugated antibodies or isotype control antibodies for 20 min at 4°C. Antibodies used were phycoerythrin-F4/80 (PE-F4/80) at 2.5 μg/mL, allophycocyanin-CD11b (APC-CD11b) at 1.25 μg/mL, and phycoerythrin-Cy7-CD11c (PE-Cy7-CD11c) at 5 μg/mL. All antibodies were obtained from eBioscience. After incubation, cells were washed twice in ice-cold PBS + 0.5% endotoxin-free BSA and resuspended in the same buffer supplemented with propidium iodide. F4/80+CD11b+ cells were separated from the remainder of the peritoneal cell population using a FACS Aria cell sorter (BD Biosciences, San Jose, CA) and collected in cooled sorting buffer. Cells were centrifuged at 500 g for 5 min at 4°C and processed for RNA extraction.
For analysis of cytokine secretion, the peritoneal lavage was enriched for CD11b+ cells using magnetic bead separation (MACS; Miltenyi Biotech). For intracellular cytokine staining, cells were plated at 5 × 105/mL and incubated at 37°C + 5% CO2 for 12 h with BD Golgi Plug (BD Biosciences). Cells were then stained using mAbs recognizing CD11b (conjugated to fluorescein isothiocyanate or PE) and CD11c (conjugated to allophycocyanin) and then stained with fluorescein isothiocyanate-TNF or PE-IL-6 (all antibodies were from BD Pharmingen, San Diego, CA). Intracellular staining was carried out according to the manufacturer's specifications using the Cytofix Cytoperm Plus kit (BD Biosciences). Specificity of staining was confirmed using isotype control antibodies. All analyses were performed on a FACSCalibur (Becton Dickinson, Franklin Lakes, NJ) using FlowJo analysis software (Treestar, Ashland, OR). For analysis of cytokine secretion, cells were incubated without Golgi plug and the supernatant was analyzed for cytokines (TNF-α, IL-6, and monocyte chemoattractant protein [MCP-1]) in the by Cytometric Bead Array Mouse Inflammation kit (BD Biosciences). Analysis was performed on FACSFortessa using FCAP Array Software (BD Biosciences).
Gene expression analysis.
Total RNA was extracted using the RNeasy mini kit (Qiagen, Valencia, CA) or TRIzol. After DNase treatment, first-strand cDNA was synthesized from template RNA using the thermoscript RT-PCR system (Invitrogen). Quantitative real time RT-PCR was then performed using Rotorgene 3000 (Corbett Research, Sydney, New South Wales, Australia) using the Assay-on-Demand gene expression kits (Applied Biosystems, Foster City, CA) as previously described (3). Relative expression levels for the genes of interest were calculated using the comparative critical threshold method after normalizing for the expression of 18S ribosomal RNA.
Protein analysis.
Tissue extracts were prepared, and immunoblotting lysates were resolved by SDS-PAGE, transferred to polyvinylidene fluoride membranes, and probed with primary antibodies for 2 h (Akt Ser473 and AMP-activated protein kinase [AMPK] T172) or overnight (Akt and AMPK-α pan) as described (3). For IRS-1 phosphorylation, lysates were immunoprecipitated with anti–IRS-1 (Upstate Biotechnology, Lake Placid, NY) and immunoblotted using the antiphosphotyrosine clone 4G10, P85 subunit of phosphatidylinositol (PI)-3-kinase or IRS-1 (Upstate Biotechnology) as described (3). Immunoblotted proteins were detected with enhanced chemiluminescence and quantified by densitometry.
Blood analysis.
Plasma insulin was measured by ELISA (Linco Research). Adipokines (leptin, TNF-α, resistin, t-plasminogen activator inhibitor 1 [tPAI-1], MCP-1) were measured with a bio-plex assay using a mouse adipokine kit (Linco Research) and manufacturer's recommendation. Liver enzymes were measured using an automated analyzer (Olympus AU 2700 chemistry immunoannalyzer).
Immunohistochemistry.
Liver tissue was collected in formalin and paraffin-embedded, or it was embedded in optimal cutting temperature compound (OCT Tissue-Tek; Sakura Fineteck, Torrance, CA) and snap-frozen. For analysis of liver morphology, 5-μm sections of formalin-fixed paraffin–embedded tissues were stained using hematoxylin and eosin. For analysis of infiltrating cell types, sequential 5-μm cryosections were acetone-fixed and stained using an avidin-biotin immunoperoxidase technique (31). Sections were stained with the following primary antibodies: F4/80 (AbD Serotec, Kidlington, Oxfordshire, U.K.); CD4, CD11c, or B220 (all from BD Pharmingen); CD8 or CD11b (both from Biolegend); followed by incubation with streptavidin DH biotinylated-horseradish peroxidase reagent (ABC Kit; VectaStain Elite; Vector Laboratories, Burlingame, CA). Sections were developed using Sigma FAST 3,3′-diaminobenzidine (Sigma) and photographed with Olympus DP Controlled (Olympus Optical).
Statistical analyses.
Data are presented as mean ± SEM. Data were analyzed using a Student t test or where appropriate a two-way ANOVA with a Bonferroni post hoc test (GraphPad PRISM, version 4.0; GraphPad Software, Inc., La Jolla, CA). P < 0.05 was considered as significant.
RESULTS
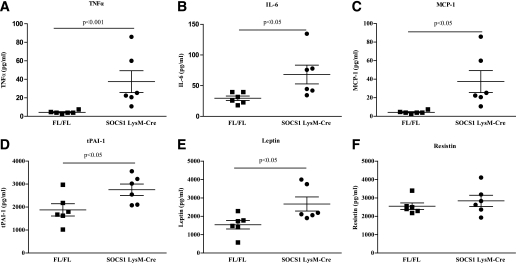
Body weight, liver, and adipose tissue weight did not differ between SOCS1 LysM-Cre mice and control littermates (Table 1). SOCS1 LysM-Cre mice had systemic inflammation as indicated by elevated plasma TNF-α (+130%), IL-6 (+560%), tPAI-1 (46%), and MCP-1 (130%) (Fig. 1A–D). Leptin levels were increased in SOCS1 LysM-Cre mice (Fig. 1E) in agreement with previous studies demonstrating stimulatory effects of TNF-α on leptin production (32). Serum resistin levels were not different between control and SOCS1 LysM-Cre mice (Fig. 1F).
TABLE 1.
Body weight, organ weights, and fasting glucose from SOCS1 LysM-Cre mice and control littermates
| Fl/FL | SOCS1 LysM-Cre | |
| Body weight (g) | 31.1 ± 1.2 | 27.4 ± 1.1 |
| Liver weight (g) | 1.45 ± 0.1 | 1.28 ± 0.1 |
| Adipose tissue weight (g) | 0.6 ± 0.2 | 0.4 ± 0.1 |
| Fasting glucose (mmol/L) | 6.1 ± 0.3 | 6.9 ± 0.2 |
Data are means ± SEM; n = 12 mice per group. For fasting glucose, P = 0.06.
FIG. 1.
Deletion of SOCS1 from macrophages results in systemic inflammation. Serum concentrations of TNF-α (A), IL-6 (B), MCP-1 (C), PAI-1 (D), leptin (E), and resistin (F) are shown. ▪, FL/FL control mice; •, SOCS1 LysM-Cre mice. Data are means ± SEM; n = 6.
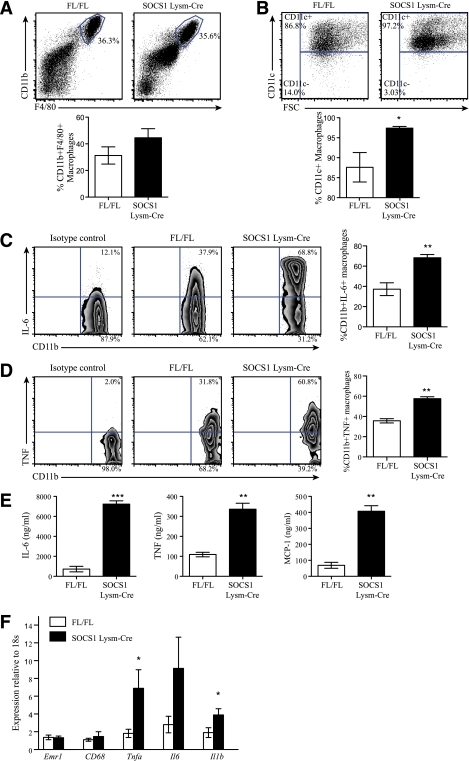
To examine whether systemic inflammation in SOCS1 LysM-Cre mice was derived from macrophages, we isolated peritoneal cells and sorted them by flow cytometry based on the expression of the macrophage markers CD11b and F4/80 and found that there were no differences in macrophage numbers between genotypes (Fig. 2A). CD11c is considered to be a critical marker of macrophage activation, and we found that, consistent with the increase in serum proinflammatory cytokines, the percentage of CD11c+ macrophages was higher in SOCS1 LysM-Cre mice (Fig. 2B). Using intracellular cytokine staining, we found ∼68% of SOCS1 LysM-Cre macrophages secreted IL-6 compared with ∼37% from littermate controls (Fig. 2C). Likewise, ∼57% SOCS1 LysM-Cre macrophages secreted TNF-α compared with ∼36% from littermate controls (Fig. 2D). We also quantitated the amount of each cytokine produced using a cytometric bead array (Fig. 2E) and found that macrophages from SOCS1 LysM-Cre mice secreted more IL-6, TNF-α, and MCP-1 compared to littermate controls. Increased inflammation was also observed in SOCS1 LysM-Cre macrophages isolated from the spleen (data not shown). The increased activation state of macrophages from SOCS1 LysM-Cre mice was also confirmed via RT-quantitative PCR analysis of bone marrow–derived macrophages (Supplementary Table 1). Collectively, these data demonstrate that macrophages from SOCS1 LysM-Cre mice are constitutively more activated compared with macrophages from control mice.
FIG. 2.
Macrophages from SOCS1 LysM-Cre mice are constitutively activated and inflamed. Peritoneal cells were isolated and stained with antibodies against F4/80, CD11b, CD11c, or isotype controls and analyzed by flow cytometry. A: Number of F4/80+CD11b+ cells in wild-type and SOCS1 LysM-Cre mice indicated as percentage of viable peritoneal cell population. B: Cells were gated for F4/80+CD11b+ cells and examined for expression of CD11c. Shown is the percentage of CD11c-positive or -negative cells within the total F4/80+CD11b+ population. A and B: Top panels are representative plots; bottom panels are means ± SEM, n = 8 per group. White bars, FL/FL control mice; black bars, SOCS1 LysM-Cre mice. Representative plots of the intracellular cytokine staining to detect secretion of IL-6 (C) and TNF-α (D) in peritoneal macrophages are shown. For each, cells were gated on the CD11b+ population, and data represented in the graph are combined from two experiments and expressed as mean ± SEM; n = 6 per group. E: Quantitation of cytokine secretion by CD11b+ peritoneal macrophages from littermate control and SOCS1 LysM-Cre mice. Secretion of IL-6, TNF, and MCP-1 by CD11b+ peritoneal macrophages was measured by cytokine bead array. F: Adipose tissue expression of Emr1, Cd68, Tnfa, Il6, and Il1b in SOCS1 LysM-Cre and control mice. Data are shown as mean ± SEM; n = 8 per group. White bars, FL/FL control mice; black bars, SOCS1 LysM-Cre mice. For all experiments, *P < 0.05, **P < 0.01, and ***P < 0.001 relative to control. (A high-quality color representation of this figure is available in the online issue.)
Given that adipose tissue macrophages are the primary source of circulating cytokines, we also examined markers of macrophage infiltration and inflammation from this tissue. Consistent with findings in the peritoneal cavity, the expression of the macrophage markers Emr1 (F4/80) and Cd68 was similar in the adipose tissue of wild-type and SOCS1 LysM-Cre mice (Fig. 2F). However, despite similar macrophage content, adipose tissue expression of Tnfa, Il6, and Il1b was increased in SOCS1 LysM-Cre mice (Fig. 2F). Taken together, these data indicate that increased systemic inflammation in SOCS1 LysM-Cre mice is associated with constitutively activated macrophages. We also have shown previously evidence for T-cell activation in these mice (23).
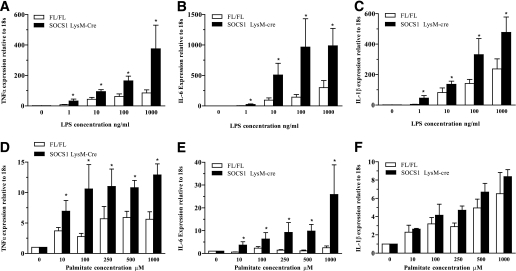
Macrophages from SOCS1 LysM-Cre mice are hypersensitive to the inflammatory effects of LPS and saturated fatty acids.
Inflammatory cytokine production after activation of TLR4 in macrophages by LPS and palmitic acid has been implicated as a potential mechanism linking inflammation to insulin resistance in obesity (33). Therefore, to test whether macrophages from SOCS1 LysM-Cre mice were hypersensitive to LPS and palmitate, we incubated bone marrow–derived macrophages from SOCS1 LysM-Cre mice and littermates with varying concentrations of these two TLR4 ligands. In the absence of the ligand, macrophage expression of Tnfa, Il6, and Il1b was low in control and SOCS1 LysM-Cre mice (Supplementary Table 1). Both LPS and palmitic acid dose-dependently increased Tnfa (Fig. 3A and D) and Il6 (Fig. 3B and E), an effect that was greater in macrophages from SOCS1 LysM-Cre mice. SOCS1 LysM-Cre macrophages also had increased Il1b mRNA in response to LPS (Fig. 3C) but not palmitate (Fig. 3F). TLR4 expression was not altered by genotype, LPS, or palmitate (data not shown). Overall, these data suggest that increased inflammation in SOCS1 LysM-Cre mice may be a result of hypersensitivity to the TLR4 ligands LPS and palmitic acid.
FIG. 3.
SOCS1 LysM-Cre mice have increased sensitivity to LPS and palmitate, Tnfa (A and D), Il6 (B and E), and Il1b (C and F) measured by RT-quantitative PCR after stimulation of bone marrow–derived macrophages from wild-type and SOCS1 LysM-Cre mice with increasing concentrations of LPS and palmitate, respectively. Data are means ± SEM; n = 3 per group. White bars, FL/FL control mice; black bars, SOCS1 LysM-Cre mice. *P < 0.05 genotype effect relative to control.
Hepatic insulin resistance in SOCS1 LysM-Cre mice.
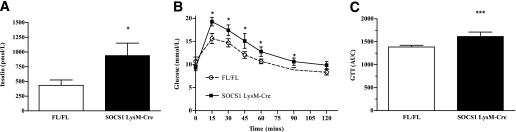
Fasting glucose (Table 1, P = 0.06) and insulin (Fig. 4A) levels were higher in SOCS1 LysM-Cre mice, relative to littermate controls. To further investigate the perturbations in glucose homeostasis of SOCS1 LysM-Cre mice, intraperitoneal glucose tolerance tests were performed, and SOCS1 LysM-Cre mice had higher glucose levels and greater area under the curve during the intraperitoneal glucose tolerance tests (Fig. 4B and C).
FIG. 4.
Glucose intolerance in SOCS1 LysM-Cre mice. A: Hyperinsulinemia and glucose intolerance (B and C) as assessed by increased area under the curve during GTT. White bars and open white circles, FL/FL control mice; black bars and black squares, SOCS1 LysM-Cre mice. Data are means ± SEM; n = 10–12. *P < 0.05 genotype effect relative to control; ***P < 0.001 relative to control.
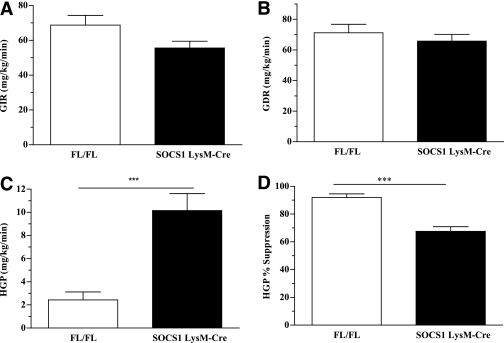
To assess the specific contribution of hepatic versus peripheral insulin resistance to the perturbed glucose homeostasis observed in SOCS1 LysM-Cre mice euglycemic-hyperinsulinemic-clamp studies were then performed. Basal glucose turnover was similar between the two groups of mice (Supplementary Table 2). The glucose infusion rate (the amount of 50% dextrose solution infused to maintain euglycemia) tended to be lower in SOCS1 LysM-Cre mice (Fig. 5A, P = 0.09). The glucose disposal rate (a measure of predominantly skeletal muscle insulin sensitivity) was not different between groups (Fig. 5B). However, hepatic glucose production (HGP) during the clamp was higher and was suppressed to a lesser degree in SOCS1 LysM-Cre mice (Fig. 5C and D). These results indicate that SOCS1 LysM-Cre mice have hepatic insulin resistance. Consistent with the similar rates of insulin-stimulated glucose disposal measured in vivo during the clamp, glucose uptake (Supplementary Fig. 1A) and Akt phosphorylation (Supplementary Fig. 1B) in isolated extensor digitorum longus muscles were not different between genotypes. This was despite greater muscle inflammation as indicated by elevated Tnfa, Il6, Il1b, and Emr1 (Supplementary Fig. 1C). To elucidate the mechanism underlying preserved muscle insulin sensitivity, despite the presence of greater muscle inflammation, we measured skeletal muscle AMPK phosphorylation (Supplementary Fig. 1D). We found skeletal muscle AMPK phosphorylation to be preserved in SOCS1 LysM-Cre mice, suggesting that IL-6 may have offset the negative effects of TNF-α on these signaling pathways in skeletal muscle (3,46).
FIG. 5.
Hepatic insulin resistance in SOCS1 LysM-Cre mice. Glucose infusion rate (GIR) (A) and glucose disposal rate (GDR) (B). HGP (C) and HGP% (D) suppression during euglycemic clamp are shown. White bars, FL/FL control mice; black bars, SOCS1 LysM-Cre mice. Data are means ± SEM; n = 10–12. ***P < 0.001 relative to control.
Reduced hepatic insulin signaling and increased liver inflammation in SOCS1 LysM-Cre mice.
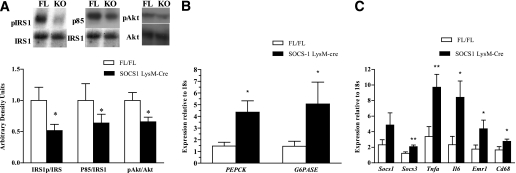
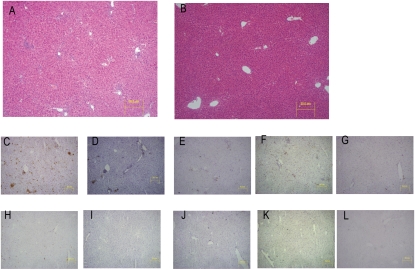
To determine the mechanism of reduced HGP suppression observed during the clamp studies, we examined insulin signaling in the liver during the clamp. IRS-1, IRS-1–associated PI3-kinase and Akt phosphorylation were all reduced in the liver of SOCS1 LysM-Cre mice after the clamp (Fig. 6A). Consistent with reduced insulin signaling and increased HGP during the clamp, the expression of Pck1 and G6pc were dramatically increased in SOCS1 LysM-Cre mice (Fig. 6B). Furthermore, the mRNA expression of Socs3, Tnfa, and Il6, as well as the macrophage markers Emr1 and Cd68, were increased in the liver of SOCS1 LysM-Cre mice (Fig. 6C). There was also a trend to increased Socs1 expression, although this did not reach statistical significance. The liver inflammation in SOCS1 LysM-Cre mice was accompanied by an increase in inflammatory cell infiltrate (Fig. 7) and liver enzymes (Supplementary Table 3). However, liver synthetic function was unaffected as highlighted by similar albumin and bilirubin levels in both genotypes (Supplementary Table 3). This suggests that liver failure was not present in SOCS1 LysM-Cre mice at the time of analysis despite the presence of transaminitis. Taken together, these data indicate that the elevated cytokines associated with macrophage accumulation, liver inflammation, and upregulation of SOCS proteins may lead to hepatic insulin resistance in SOCS1 LysM-Cre mice.
FIG. 6.
Impaired hepatic insulin signaling in SOCS1 LysM-Cre mice is associated with increased liver inflammation. Reduced IRS-1 tyrosine phosphorylation, IRS-1–associated p85-subunit of PI-3 kinase, and Akt phosphorylation in liver of SOCS1 LysM-Cre mice (A) are shown. B: Increased expression of PEPCK and G6Pase in clamped liver as measured via RT-quantitative PCR. C: Increased inflammatory signaling in liver of SOCS1 LysM-Cre mice. Data are means ± SEM; n = 12 mice per group. White bars, FL/FL control mice; black bars, SOCS1 LysM-Cre mice. *P < 0.05 and **P < 0.01 relative to control. KO, SOCS1 LysM-Cre.
FIG. 7.
Increased liver inflammatory cell infiltration in SOCS1 LysM-Cre mice. Liver sections from SOCS1 LysM-Cre (A and C–G) and FL/FL controls (B and H–L) stained as follows: hematoxylin-eosin (A and B), CD11b (C and H), CD11c (D and I), CD4 (E and J), CD8 (F and K), and B220 (G and L). (A high-quality digital representation of this figure is available in the online issue.)
DISCUSSION
Obesity-related insulin resistance is a multifaceted disease involving defects in multiple systems, and the subclinical release of inflammatory mediators by macrophages is one of many factors (3–5). However, the mechanisms underlying the overactivation of the innate immune system in obesity are not fully understood. In the current study, using mice that have constitutively activated macrophages (SOCS1 LysM-Cre mice), we demonstrate the importance of macrophage SOCS1 in maintaining hepatic insulin sensitivity. We show that SOCS1 LysM-Cre mice develop hepatic insulin resistance even in the absence of other factors contributing to obesity-induced insulin resistance. These findings suggest that therapies aimed at reducing SOCS1 in cells indiscriminately may increase systemic inflammation and be detrimental to insulin action.
The SOCS-1 LysM-Cre mouse is a model of inflammatory disease that occurs independent of changes in fat mass/obesity. In adipose tissue of obese humans and rodents, there is an increase in both the content as well as activation of adipose tissue macrophages (4,5,13,34). Although SOCS1 LysM-Cre mice have dramatic increases in macrophage activation, there was no increase in adipose tissue macrophage number. These data demonstrate that inflammation in itself is not sufficient to drive adipose tissue macrophage recruitment despite increases in circulating cytokines such as MCP-1, suggesting that additional factors related to the enlarged adipocyte, such as hypoxia (35) or free fatty acids (36), are important for adipose tissue macrophage recruitment in obesity.
Mice with hematopoietic deletion of TLR4 have reduced obesity-related inflammation and insulin resistance (37). The TLR4 ligands LPS and saturated fatty acids are both elevated in obesity, and this may explain the beneficial effects of TLR4 deletion on these parameters. Although it has been recognized for over a decade that SOCS1 is an important regulator of immune and inflammatory responses (22,23), more recent studies have demonstrated that the mechanisms by which SOCS1 inhibits inflammation involve downregulation of TLR4 signaling (21). In the current study, we demonstrate that SOCS1 LysM-Cre mice have increased CD11c+ macrophages and elevated circulating levels of inflammatory cytokines. In agreement with previous studies, we show that an important source of this increased inflammatory response is not only heightened sensitivity to LPS (current study and 23,38) but also to the saturated fatty acid palmitate. Future studies in SOCS1 LysM-Cre mice treated with antibiotics or nicotinic acid, to reduce circulating LPS and free fatty acids, respectively, are warranted to confirm the importance of the SOCS1/TLR4 pathway in vivo.
In the current study euglycemic-hyperinsulinemic clamps showed that the hyperinsulinemia and glucose intolerance of SOCS1 LysM-Cre mice was a result of insulin resistance in the liver. Consistent with this, we found that IRS-1 phosphorylation, IRS-1 associated p85 subunit of PI-3 kinase, and Akt phosphorylation were all reduced in the liver of SOCS1 LysM-Cre mice. Furthermore, this insulin resistance was associated with increased Socs3 expression consistent with previous reports showing an essential role for SOCS3 in mediating cytokine-induced liver insulin resistance (28). The exact cause of hepatic insulin resistance in SOCS1 LysM-Cre mice is not known since we observed increases in both systemic and local (liver) inflammation. The observed increases in circulating cytokines such as TNF-α, IL-6, and IL-1β were higher than typically observed in obesity; however, our observed increases in liver macrophage content and activation are consistent with other rodent models of obesity (28,39,40). In support of the critical role of macrophages in mediating the liver insulin resistance of SOCS1 LysM-Cre mice, we have found that liver-specific deletion of SOCS1 does not affect serum glucose or insulin levels, glucose or insulin tolerance or hepatic insulin sensitivity as assessed by euglycemic-hyperinsulinemic clamp (N.S., S.L.F., G.R.S., and T.W.K., unpublished observations).
Our findings do pose the interesting question of why inflammation in SOCS1 LysM-Cre mice did not cause muscle insulin resistance. One possibility may be because of the elevated levels of IL-6 in SOCS1 LysM-Cre mice. Chronic elevations in IL-6 promote whole-body insulin sensitivity in lean (41) or obese mice (42). Consistent with this IL-6 increases skeletal muscle glucose disposal and fatty acid oxidation (43,44), effects that may be mediated through increases in Akt and AMPK. In contrast, TNF-α–induced skeletal muscle insulin resistance in part involves suppression of skeletal muscle AMPK signaling (3). However, in the current study, we found no difference in skeletal muscle Akt (Supplementary Fig. 1B) or AMPK (Supplementary Fig. 1D) phosphorylation, which may be because IL-6 had offset the negative effects of TNF-α on these signaling pathways. Although the reasons behind the tissue-specific effects of IL-6 on insulin action remain unknown, it has been speculated that low density of the signaling gp130 receptor in skeletal muscle (45), which results in reduced SOCS3 expression, may be important (46). Consistent with this, IL-6 induces SOCS3 expression by twofold in muscle but 25-fold in liver (46). Therefore, although speculative, increased IL-6 levels may have negated the detrimental effects of TNF-α on skeletal muscle insulin action in SOCS1 LysM-Cre mice. Future studies following IL-6 neutralization in SOCS1 LysM-Cre mice may help determine whether this is an important factor.
Genetic polymorphisms in the promoter regions of the SOCS1 gene that increase its transcriptional activity have been shown to associate with insulin sensitivity despite the presence of obesity and higher BMI (24). Because SOCS1 directly inhibits insulin signaling through ubiquitination of IRS-2 (26,27), the finding that increased transcriptional activity of SOCS1 is associated with improved insulin sensitivity was difficult to comprehend. However, in light of our findings demonstrating the pronounced hyperinsulinemia, glucose intolerance, and hepatic insulin resistance in SOCS1 LysM-Cre mice, it is possible that attenuation of macrophage inflammation may be the primary mechanism by which this polymorphism, which increases SOCS1 expression, improves insulin sensitivity (24). Taken together, these data suggest that macrophage expression of SOCS1 is a critical regulator of inflammation and hepatic insulin sensitivity and that strategies aimed at increasing macrophage SOCS1 expression may be important for the treatment of insulin resistance.
Supplementary Material
Acknowledgments
This work was supported by grants and fellowships from the National Health and Medical Research Council (NHMRC) of Australia (T.W.K. and G.R.S.) and the Canadian Institutes of Health Research (G.R.S.).
G.R.S. is a Canadian Research Chair in Metabolism and Obesity. N.S. is supported by a postgraduate research scholarship. K.L.G. is supported by fellowships from the Australian Diabetes Society and the Juvenile Diabetes Research Foundation.
No potential conflicts of interest relevant to this article were reported.
N.S. researched data, contributed to the discussion, and wrote the manuscript. K.L.G. and S.G. researched data, contributed to the discussion, and reviewed and edited the manuscript. J.E.H. researched data and contributed to the discussion. S.L.F. and K.A.H. assisted with experiments. G.R.S. and T.W.K. designed the study, contributed to the discussion, and edited the manuscript.
The authors thank Dr. Prithi Bhathal, experienced liver histopathologist, Melbourne Pathology, Australia, for reviewing the liver histology.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-0259/-/DC1.
REFERENCES
- 1.Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest 2006;116:1793–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science 1993;259:87–91 [DOI] [PubMed] [Google Scholar]
- 3.Steinberg GR, Michell BJ, van Denderen BJ, et al. Tumor necrosis factor alpha-induced skeletal muscle insulin resistance involves suppression of AMP-kinase signaling. Cell Metab 2006;4:465–474 [DOI] [PubMed] [Google Scholar]
- 4.Xu H, Barnes GT, Yang Q, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest 2003;112:1821–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest 2003;112:1796–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol 2001;1:135–145 [DOI] [PubMed] [Google Scholar]
- 7.O'Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol 2007;7:353–364 [DOI] [PubMed] [Google Scholar]
- 8.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature 2006;444:1022–1023 [DOI] [PubMed] [Google Scholar]
- 9.Brun P, Castagliuolo I, Di Leo V, et al. Increased intestinal permeability in obese mice: new evidence in the pathogenesis of nonalcoholic steatohepatitis. Am J Physiol Gastrointest Liver Physiol 2007;292:G518–G525 [DOI] [PubMed] [Google Scholar]
- 10.Membrez M, Blancher F, Jaquet M, et al. Gut microbiota modulation with norfloxacin and ampicillin enhances glucose tolerance in mice. FASEB J 2008;22:2416–2426 [DOI] [PubMed] [Google Scholar]
- 11.Shi H, Kokoeva MV, Inouye K, Tzameli I, Yin H, Flier JS. TLR4 links innate immunity and fatty acid-induced insulin resistance. J Clin Invest 2006;116:3015–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsukumo DM, Carvalho-Filho MA, Carvalheira JB, et al. Loss-of-function mutation in Toll-like receptor 4 prevents diet-induced obesity and insulin resistance. Diabetes 2007;56:1986–1998 [DOI] [PubMed] [Google Scholar]
- 13.Nguyen MT, Favelyukis S, Nguyen AK, et al. A subpopulation of macrophages infiltrates hypertrophic adipose tissue and is activated by free fatty acids via Toll-like receptors 2 and 4 and JNK-dependent pathways. J Biol Chem 2007;282:35279–35292 [DOI] [PubMed] [Google Scholar]
- 14.Ghanim H, Aljada A, Hofmeyer D, Syed T, Mohanty P, Dandona P. Circulating mononuclear cells in the obese are in a proinflammatory state. Circulation 2004;110:1564–1571 [DOI] [PubMed] [Google Scholar]
- 15.Boden G, She P, Mozzoli M, et al. Free fatty acids produce insulin resistance and activate the proinflammatory nuclear factor-κB pathway in rat liver. Diabetes 2005;54:3458–3465 [DOI] [PubMed] [Google Scholar]
- 16.Nguyen MT, Satoh H, Favelyukis S, et al. JNK and tumor necrosis factor-alpha mediate free fatty acid-induced insulin resistance in 3T3-L1 adipocytes. J Biol Chem 2005;280:35361–35371 [DOI] [PubMed] [Google Scholar]
- 17.Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol 2007;7:454–465 [DOI] [PubMed] [Google Scholar]
- 18.Sakamoto H, Yasukawa H, Masuhara M, et al. A Janus kinase inhibitor, JAB, is an interferon-gamma-inducible gene and confers resistance to interferons. Blood 1998;92:1668–1676 [PubMed] [Google Scholar]
- 19.Sporri B, Kovanen PE, Sasaki A, Yoshimura A, Leonard WJ. JAB/SOCS1/SSI-1 is an interleukin-2-induced inhibitor of IL-2 signaling. Blood 2001;97:221–226 [DOI] [PubMed] [Google Scholar]
- 20.Naka T, Narazaki M, Hirata M, et al. Structure and function of a new STAT-induced STAT inhibitor. Nature 1997;387:924–929 [DOI] [PubMed] [Google Scholar]
- 21.Mansell A, Smith R, Doyle SL, et al. Suppressor of cytokine signaling 1 negatively regulates Toll-like receptor signaling by mediating Mal degradation. Nat Immunol 2006;7:148–155 [DOI] [PubMed] [Google Scholar]
- 22.Starr R, Metcalf D, Elefanty AG, et al. Liver degeneration and lymphoid deficiencies in mice lacking suppressor of cytokine signaling-1. Proc Natl Acad Sci USA 1998;95:14395–14399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chong MM, Metcalf D, Jamieson E, Alexander WS, Kay TW. Suppressor of cytokine signaling-1 in T cells and macrophages is critical for preventing lethal inflammation. Blood 2005;106:1668–1675 [DOI] [PubMed] [Google Scholar]
- 24.Gylvin T, Ek J, Nolsøe R, et al. Functional SOCS1 polymorphisms are associated with variation in obesity in whites. Diabetes Obes Metab 2009;11:196–203 [DOI] [PubMed] [Google Scholar]
- 25.Harada M, Nakashima K, Hirota T, et al. Functional polymorphism in the suppressor of cytokine signaling 1 gene associated with adult asthma. Am J Respir Cell Mol Biol 2007;36:491–496 [DOI] [PubMed] [Google Scholar]
- 26.Ueki K, Kondo T, Kahn CR. Suppressor of cytokine signaling 1 (SOCS-1) and SOCS-3 cause insulin resistance through inhibition of tyrosine phosphorylation of insulin receptor substrate proteins by discrete mechanisms. Mol Cell Biol 2004;24:5434–5446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jamieson E, Chong MM, Steinberg GR, et al. Socs1 deficiency enhances hepatic insulin signaling. J Biol Chem 2005;280:31516–31521 [DOI] [PubMed] [Google Scholar]
- 28.Sachithanandan N, Fam BC, Fynch S, et al. Liver-specific suppressor of cytokine signaling-3 deletion in mice enhances hepatic insulin sensitivity and lipogenesis resulting in fatty liver and obesity. Hepatology 2010;52:1632–1642 [DOI] [PubMed] [Google Scholar]
- 29.Dzamko N, van Denderen BJ, Hevener AL, et al. AMPK beta1 deletion reduces appetite, preventing obesity and hepatic insulin resistance. J Biol Chem 2010;285:115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinberg GR, Watt MJ, Ernst M, Birnbaum MJ, Kemp BE, Jørgensen SB. Ciliary neurotrophic factor stimulates muscle glucose uptake by a PI3-kinase-dependent pathway that is impaired with obesity. Diabetes 2009;58:829–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas HE, Parker JL, Schreiber RD, Kay TW. IFN-gamma action on pancreatic beta cells causes class I MHC upregulation but not diabetes. J Clin Invest 1998;102:1249–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirchgessner TG, Uysal KT, Wiesbrock SM, Marino MW, Hotamisligil GS. Tumor necrosis factor-alpha contributes to obesity-related hyperleptinemia by regulating leptin release from adipocytes. J Clin Invest 1997;100:2777–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JY, Sohn KH, Rhee SH, Hwang D. Saturated fatty acids, but not unsaturated fatty acids, induce the expression of cyclooxygenase-2 mediated through Toll-like receptor 4. J Biol Chem 2001;276:16683–16689 [DOI] [PubMed] [Google Scholar]
- 34.Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest 2007;117:175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye J, Gao Z, Yin J, He Q. Hypoxia is a potential risk factor for chronic inflammation and adiponectin reduction in adipose tissue of ob/ob and dietary obese mice. Am J Physiol Endocrinol Metab 2007;293:E1118–E1128 [DOI] [PubMed] [Google Scholar]
- 36.Kosteli A, Sugaru E, Haemmerle G, et al. Weight loss and lipolysis promote a dynamic immune response in murine adipose tissue. J Clin Invest 2010;120:3466–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saberi M, Woods NB, de Luca C, et al. Hematopoietic cell-specific deletion of toll-like receptor 4 ameliorates hepatic and adipose tissue insulin resistance in high-fat-fed mice. Cell Metab 2009;10:419–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kinjyo I, Hanada T, Inagaki-Ohara K, et al. SOCS1/JAB is a negative regulator of LPS-induced macrophage activation. Immunity 2002;17:583–591 [DOI] [PubMed] [Google Scholar]
- 39.Huang W, Metlakunta A, Dedousis N, et al. Depletion of liver Kupffer cells prevents the development of diet-induced hepatic steatosis and insulin resistance. Diabetes 2010;59:347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Odegaard JI, Ricardo-Gonzalez RR, Red Eagle A, et al. Alternative M2 activation of Kupffer cells by PPARdelta ameliorates obesity-induced insulin resistance. Cell Metab 2008;7:496–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holmes AG, Mesa JL, Neill BA, et al. Prolonged interleukin-6 administration enhances glucose tolerance and increases skeletal muscle PPARalpha and UCP2 expression in rats. J Endocrinol 2008;198:367–374 [DOI] [PubMed] [Google Scholar]
- 42.Sadagurski M, Norquay L, Farhang J, D'Aquino K, Copps K, White MF. Human IL6 enhances leptin action in mice. Diabetologia 2010;53:525–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petersen EW, Carey AL, Sacchetti M, et al. Acute IL-6 treatment increases fatty acid turnover in elderly humans in vivo and in tissue culture in vitro. Am J Physiol Endocrinol Metab 2005;288:E155–E162 [DOI] [PubMed] [Google Scholar]
- 44.Carey AL, Steinberg GR, Macaulay SL, et al. Interleukin-6 increases insulin-stimulated glucose disposal in humans and glucose uptake and fatty acid oxidation in vitro via AMP-activated protein kinase. Diabetes 2006;55:2688–2697 [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Pilon G, Marette A, Baracos VE. Cytokines and endotoxin induce cytokine receptors in skeletal muscle. Am J Physiol Endocrinol Metab 2000;279:E196–E205 [DOI] [PubMed] [Google Scholar]
- 46.Weigert C, Hennige AM, Lehmann R, et al. Direct cross-talk of interleukin-6 and insulin signal transduction via insulin receptor substrate-1 in skeletal muscle cells. J Biol Chem 2006;281:7060–7067 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.