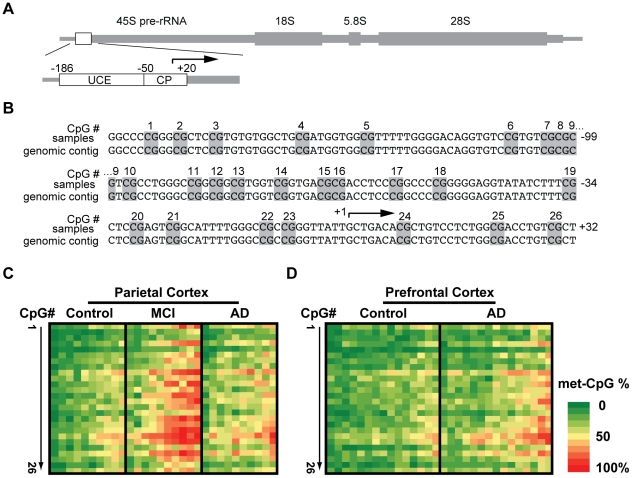
Figure 1. Methyl-cytosine (mC) mapping of the rDNA promoter region.
A, The rDNA gene unit. The location of the promoter region that was selected for bisulfite mapping of methyl cytosines is indicated by a box. That analyzed region included the Upstream Control Element (UCE) and the Core Promoter (CP) which contain 26 CpG methylation sites that are critical for epigenetic regulation of nucleolar transcription (for more detail, see Fig. S1). B. Nucleotide sequence of the human rDNA promoter. The numbers on the right indicate positions relative to the transcription start site at position +1 (arrow); Potentially methylated CpG dinucleotides are marked grey and numbered. C, Bisulfite sequencing analysis of parietal cortex from control-, MCI-, and AD individuals (n = 10 for each group). D. Bisulfite sequencing analysis of prefrontal cortex from control-, AD individuals (n = 15 for each group). In C–D, each column represents one individual. Rows represent each of the potentially methylated 26 CpG sites in the rDNA promoter in 5′ to 3′ orientation. The data (% methylation for a given CpG) are from analyzing 20 unique clones for each subject.