Summary
It was reported previously that the major fraction of the recent decrease of tuberculosis incident cases in Arkansas had been due to a decrease in the reactivated infections. Preventing transmission of Mycobacterium tuberculosis is the key to a continued decline in tuberculosis cases. In this study, we integrated epidemiological data analysis and comparative genomics to identify host and microbial factors important to tuberculosis transmission. A significantly higher proportion of cases in large clusters (containing >10 cases) were non-Hispanic black, homeless, less than 65 years old, male sex, smear-positive sputum, excessive use of alcohol, and HIV sero-positive, compared to cases in small clusters (containing 2–5 cases) diagnosed within one year. However, being non-Hispanic black and homeless within the past year were the only two host characteristics that were identified as independent risk factors for being in large clusters. This finding suggests that social behavioral factors have a more important role in transmission of tuberculosis than does the infectiousness of the source. Comparing the genomic content of one of the large cluster strains to that of a non-clustered strain from the same community identified 25 genes that differed between the two strains, potentially contributing to the observed differences in transmission.
Introduction
Following a peak in tuberculosis cases in the United States in 1992, the incidence of tuberculosis has decreased approximately by half, with 11,540cases reported in 2009 [1]. Although the tuberculosis incidence rate in 2009 was reported by the CDC to be the lowest recorded since 1953 when the national tuberculosis reporting began, the annual percentage decline in tuberculosis incidence rate has slowed from 7.3% per year during 1993–2000 to 3.8% during 2000–2007 [1–5]. Furthermore, tuberculosis incidence rates vary greatly among the states. Arkansas, which once had incidence rates above the national average, has had a 60% decrease in tuberculosis cases from 1992 to 2006, and the incidence rate for the state has now been below the national average for the past seven years [1–6]. A study by France et al. used DNA genotyping of Mycobacterium tuberculosis isolates from Arkansas to distinguish between cases of tuberculosis resulting from recent transmission (clusters of isolates having similar DNA genotyping patterns) and cases resulting from reactivation of a latent infection acquired in the past. By examining the changes in the incidence of recently transmitted disease and reactivation disease, it was observed that the overall decline in tuberculosis cases in Arkansas resulted primarily from declining rates of reactivation disease, and less so from declining rates from recent transmission [7].
Previous studies have identified risk factors for clustering including male gender, non-Hispanic black race, younger age, homelessness, alcohol use, or intravenous drug use within the past 12 months, HIV-positivity, cavitary disease, and sputum smear positivity [8–11]. More recent studies have focused on examining host factors that are predictive of large clusters, which represent either more extensive transmission of the strain or a higher proportion of primary tuberculosis among those infected with the strain. A study by Kik et al. compared the host characteristics of the first two cases in a cluster between small (2–4 cases) and large (≥ 5 cases) clusters and found that a short interval (< 3 months) between the diagnosis of the first two patients, age < 35 years, urban residence, and sub-Saharan African nationality were independent predictors of large clusters [12].
Preventing transmission of M. tuberculosis is the key to a continued decline in tuberculosis cases. A better understanding of the host, environmental, and bacterial factors that are associated with clustering will inform strategies to prevent M. tuberculosis transmission. This study investigated host risk factors for belonging to a cluster of tuberculosis cases as well as belonging to a large cluster, and also examined bacterial factors involved in clustering by comparing the genetic content of a large cluster strain to that of a non-clustered strain from the same community.
Material and Methods
Study population
The study population included 993 tuberculosis cases having M. tuberculosis isolates collected between January 1, 1996 and December 31, 2003, representing 70.8% of all incident cases of tuberculosis and 96.9 % of the bacteriologically-confirmed cases during that time period. Patient information for each case was collected using the CDC Report of Verified Case of Tuberculosis (RVCT) form. The data collected included patient demographics (sex, race/ethnicity, age, city and county of residence, and homelessness or excessive alcohol use within the past year), clinical characteristics (site of disease, chest radiograph results, sputum culture and smear results, and HIV co-infection), and treatment regimen. The study protocols and procedures for the protection of human subjects were approved by the Health Sciences Institutional Review Boards of the University of Michigan and the University of Arkansas for Medical Sciences.
Classification of study subjects and isolates
These cases had been previously grouped into clustered or non-clustered based on IS6110 fingerprinting and spoligotyping of the isolates [7]. For isolates having six or more IS6110 copies, isolates with identical IS6110 fingerprints or isolates with IS6110 fingerprints differing by one band and having identical spoligotype patterns were designated as clustered [13]. For isolates with less than six IS6110 copies, isolates having identical IS6110 fingerprints and spoligotypes were designated as clustered. To increase the specificity of genotypic clustering as a measure for recent transmission, cases having matching genotypes by the above criteria also had to have been diagnosed within one year of each other to be considered clustered [7]. By using these criteria, 392 cases shared identical or highly similar genotype patterns with another isolate collected from the Arkansas population during the same time period.
Determination of host risk factors for clustering
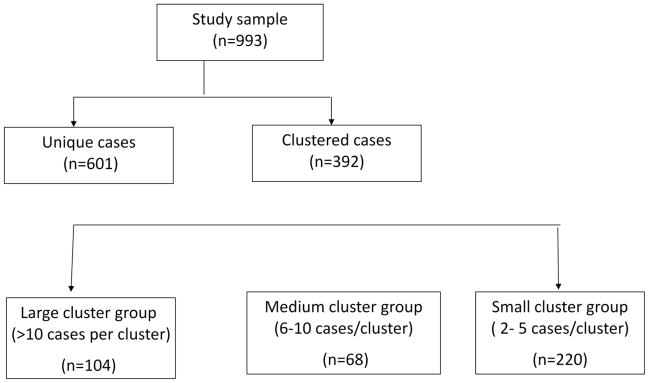
To identify host risk factors for clustering of tuberculosis cases in the Arkansas population, the distribution of previously identified host risk factors for TB transmission [8–11] was first compared between the 392 clustered cases and the 601 non-clustered cases by chi-square or Fisher’s exact test, as appropriate. The host risk factors analyzed were non-Hispanic black race, male sex, age less than 65 years, homelessness, alcohol use, or intravenous drug use within the past 12 months, HIV-positive, cavitary disease, and sputum smear positivity. To identify host risk factors for large clusters of tuberculosis cases, the clustered cases were divided into three groups (Figure 1): large cluster cases (containing >10 cases), medium cluster cases (containing 6–10 cases), and small cluster cases (2–5 cases). The distribution of host demographic characteristics was then compared between the small and the medium cluster groups, and between the small and the large cluster groups, respectively by chi-square or Fisher’s exact test, as appropriate. In order to identify the host risk factors for clusters of various size ranges, respectively, controlling for potential confounders, two multivariate logistical regression models were fit, using the small cluster group as a control group. Variables included in these two models are essential demographic variables, such as age, sex, and race/ethnicity, and all the other variables that had shown disproportional distributions in the Chi-square analysis. All the statistical analysis were done using SAS version 9.2. (SAS Institute, Cary, NC).
Figure 1.
Sample grouping and criteria for categorization of clusters Talarico et.al., Figure 1.
Selection of isolates for genomic comparison
Two M. tuberculosis isolates were selected for genomic comparison to identify large genomic deletions that may account for the observed differences in transmissibility between strains of M. tuberculosis. One isolate (SA201) was selected to represent the strain responsible for the large, persisting cluster of tuberculosis cases. Another isolate (SA178) that caused disease in only one person in the same setting and time period was selected for the comparison.
The criteria for selection of the comparison strain included both clinical and demographic information to assure that the unique strain’s opportunity for transmission was comparable to that of the clustered strain. First, isolates with unique IS6110 fingerprinting patterns from the counties that also had cases caused by the SA201 strain were selected as potential comparison isolates. The clinical information for the unique isolates was reviewed and two strains that were isolated from patients with pulmonary cavitary tuberculosis and were sputum smear positive were identified. From these two strains, the one that was isolated from the patient who was younger at the time of diagnosis was selected for the comparison. The patient infected with M. tuberculosis strain SA178 was 66 years old at the time of her diagnosis in 1996 and resided in the same county for her entire life. Contact investigation of this patient identified 17 contacts, all of whom were tuberculin skin tested negative, all showing 0 mm induration except for one.
Comparison of genomic content
A microarray-based genomic characterization was performed to identify large sequence polymorphisms (LSPs) in the genome of the isolate (SA201) causing the persistent cluster and the non-clustered isolate (SA178). Single channel DNA microarray hybridizations using the TIGR M. tuberculosis microarray were performed in duplicate. The microarray contains 4,750 70-mer oligonucleotides, printed twice on each slide, representing 4,127 open reading frames (ORFs) from H37Rv and 623 unique ORFs from CDC1551. Four μg of genomic DNA from each strain was digested with the restriction enzyme RsaI. Two μg of the purified digested DNA was labeled with Fluorescein-12-dCTP (PerkinElmer, Boston, MA) using the BioPrime Labeling Kit according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). A hybridization mixture of 48 μl of 1.25X HybIt hybridization buffer (ArrayIt, TeleChem International, Sunnyvale, CA), 0.6 μl of 10 mg/ml salmon sperm DNA (Invitrogen, Carlsbad, CA), and concentrated probe was prepared. The hybridization mixture was denatured at 94°C for 4 minutes before being applied to the microarray slide. The slide was then transferred to a sealed hybridization chamber and submerged in a 68°C water bath for at least 16 hours. Following hybridization, the slide was washed in 50°C low stringency wash buffer (1X SSC, 0.2% SDS) for 8 minutes. The slide was then washed in high stringency wash buffer (0.1X SSC, 0.2% SDS) for 8 minutes followed by two washes in 0.1X SSC for three minutes. Detection of hybridized fluoresce-labeled DNA probe was performed using the MICROMAX TSA Labeling and Detection Kit (PerkinElmer, Boston, MA) according to the manufacturer’s instructions. The hybridized microarray slides were scanned and Cyanine 3 intensities were analyzed using the Virtek Chip Reader (Waterloo, Ontario, Canada). Spot signal intensities that were greater than three times background intensity were counted as positive for hybridization. Each ORF was tested for hybridization at four spots for each strain. ORFs that were negative for hybridization at two, three, or four of the four spots were considered as possibly having large-sequence polymorphisms (LSPs).
To confirm the potential LSPs and determine their location and size, PCR amplification of the ORFs identified as having a potential LSP was performed, followed by automated DNA sequencing of the amplification product. Sequencing was performed in Applied Biosystems DNA Sequencers (Models 3700 and 3730). The gene sequences were compared to that of the appropriate sequenced M. tuberculosis strain (H37Rv or CDC1551) using the BLAST program of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/BLAST).
RESULTS
Host risk factors for clustering
Several demographic characteristics of the study subjects were found to be differentially distributed between all cases clustered within 1 year and non-clustered cases (Table 1). Host factors that were significantly over-represented among the clustered cases were age less than 65 years, non-Hispanic black race/ethnicity, homelessness, excessive alcohol use, being HIV-positive, and sputum smear-positive. The distribution of the host factors male sex, intravenous drug use, and lung cavitation were not significantly different between the two groups. After controlling for the potential confounders by multivariate logistical regression analysis, being non-Hispanic black race/ethnicity [OR=2.07, 95% CI (1.52, 2.82), P<0.0001] and younger than 65 years [OR=2.30, 95% CI (1.68, 3.14), P<0.0001], remained statistically significantly associated with clustering (Table 2).
Table 1.
Distribution of demographic characteristics and previously-known risk factors of TB among 392 clustered tuberculosis cases and 601 non-clustered tuberculosis cases diagnosed in Arkansas between 1996 and 2003
| Variables | clustered cases
|
non-clustered cases
|
P |
|---|---|---|---|
| N (%) | N (%) | ||
| Age | <0.0001 | ||
| <65 years | 264 (67.4) | 262 (43.6) | |
| ≥ 65 years | 128 (32.7) | 339 (56.4) | |
| Sex | 0.058 | ||
| Male | 267 (68.1) | 374 (62.2) | |
| Female | 125 (31.9) | 227 (37.8) | |
| Race | <0.0001 | ||
| Non-Hispanic black | 189 (48.2) | 167 (28.0) | |
| Other | 203 (51.8) | 430 (72.0) | |
| Homeless within past year | 0.003 | ||
| Yes | 17 (4.4) | 8 (1.3) | |
| No | 370 (95.6) | 589 (98.7) | |
| Excessive alcohol use within past year | <0.0001 a | ||
| Yes | 77 (19.8) | 51 (8.6) | |
| No | 300 (77.1) | 526 (88.4) | |
| Unknown | 12 (3.1) | 18 (3.0) | |
| Intravenous drug use within past year | 0.319* | ||
| Yes | 2 (0.5) | 1 (0.2) | |
| No | 252 (64.3) | 407 (67.7) | |
| Unknown | 138 (35.2) | 193 (32.1) | |
| HIV status | <0.0001 b | ||
| Sero - positive | 18 (4.6) | 24 (4.0) | |
| Sero - negative | 242 (61.7) | 268 (44.6) | |
| Unknown | 132 (33.7) | 309 (51.4) | |
| Lung cavitation | 0.065 | ||
| Yes | 132 (33.7) | 170 (28.3) | |
| No | 201 (51.3) | 311 (51.7) | |
| Unknown | 59 (15.0) | 120 (20.0) | |
| Sputum smear | 0.024 c | ||
| Positive | 156 (39.8) | 191 (31.8) | |
| Negative | 195 (49.7) | 327 (54.4) | |
| Unknown | 41 (10.5) | 83 (13.8) |
P-value for variable excessive alcohol use without unknowns is <0.0001.
P-value for variable HIV status without unknowns is 0.57.
P-value for variable sputum smear status without unknowns is 0.03.
P-value for Fisher’s exact test
Table 2.
Multivariate logistic regression analysis determining risk factors for being in clusters among 993 bacteriologically-confirmed tuberculosis cases diagnosed in Arkansas between 1996 and 2003
| Variables | OR | 95% CI | P |
|---|---|---|---|
| Age | <.0001 | ||
| ≥65 years | 1.00 | referent | |
| <65 years | 2.30 | 1.68–3.14 | |
| Sex | 0.32 | ||
| Male | 1.00 | referent | |
| Female | 0.85 | 0.63–1.17 | |
| Race | <.0001 | ||
| Other | 1.00 | referent | |
| non-Hispanic black | 2.07 | 1.52–2.82 | |
| Homeless within past year | 0.1 | ||
| No | 1.00 | referent | |
| Yes | 2.17 | 0.87–5.43 | |
| Excessive alcohol use within past year | 0.17 | ||
| No | 1.00 | referent | |
| Yes | 1.36 | 0.88–2.10 | |
| Sputum smear | 0.29 | ||
| Negative | 1.00 | referent | |
| Positive | 1.18 | 0.87–1.59 |
To examine host factors for being involved in large clusters, which could be the result of increased transmission and/or a higher likelihood of recent development of TB disease, the 392 clustered cases were divided into three groups based on cluster size, as described above. One hundred and four isolates contained in 5 clusters were grouped into the large cluster group, 68 isolates contained in 9 clusters were grouped into the medium cluster group, and the remaining 220 clustered isolates representing 91 clusters were grouped into the small cluster group. Host factors that were significantly over-represented among the large cluster cases, as compared with small cluster cases, were age less than 65 years, male sex, non-Hispanic black race/ethnicity, homelessness, and excessive alcohol use, HIVsero-positive, and sputum smear-positive (Table 3). The distribution of the host factors cavitary disease, and intravenous drug use were not significantly different between large cluster cases and small cluster cases. After controlling for the potential confounders by multivariate logistical regression analysis, being non-Hispanic black race/ethnicity (OR=1.89, 95% CI [1.01, 3.55], P=0.047) and homeless (OR=10.35, 95% CI [2.02, 52.93], P=0.005) remained statistically significantly associated with being in large clusters (Table 4). HIV was the only host factor that was significantly overrepresented among the medium cluster cases, in comparison with small cluster cases (Table 3). It was also the only independent risk factor for being in medium clusters (Table 4).
Table 3.
Distribution of demographic characteristics and previously known risk factors of TB among 220 small cluster tuberculosis cases, 68 medium cluster cases and 104 large cluster cases diagnosed in Arkansas between 1996 and 2003
| Classification of Clusters | |||||
|---|---|---|---|---|---|
| Variables | Small
|
Medium
|
P€ | Large
|
p¥ |
| N (%) | N (%) | N (%) | |||
| Age | 0.15 | 0.014 | |||
| <65 years | 144 (65.5) | 38 (55.9) | 82 (78.9) | ||
| ≥ 65 years | 76 (34.6) | 30 (44.1) | 22 (21.2) | ||
| Sex | 0.86 | 0.047 | |||
| Male | 143 (65.0) | 45 (66.2) | 79 (76.0) | ||
| Female | 77 (35.0) | 23 (33.8) | 25 (24.0) | ||
| Race | 0.93 | 0.049 | |||
| Non-Hispanic black | 99 (45.0) | 31 (45.6) | 59 (56.7) | ||
| Other | 121 (55.0) | 37 (54.4) | 45 (43.3) | ||
| Homeless within past year | 0.12* | <0.0001* | |||
| Yes | 3 (1.4) | 3 (4.4) | 11 (10.6) | ||
| No | 212 (98.6) | 65 (95.6) | 93 (89.4) | ||
| Excessive alcohol use within past year | 0.43 | 0.017b | |||
| Yes | 36 (16.4) | 10 (14.7) | 31 (29.8) | ||
| No | 176 (80.0) | 53 (77.9) | 71 (68.3) | ||
| Unknown | 8 (3.6) | 5 (7.4) | 2 (1.9) | ||
| Intravenous drug use within past year | 0.09* | 0.662* | |||
| Yes | 1 (0.5) | 0 (0.0) | 1 (1.0) | ||
| No | 135 (61.4) | 51 (75.0) | 66 (63.5) | ||
| Unknown | 84 (38.2) | 17 (25.0) | 37 (35.6) | ||
| HIV status | 0.04*a | 0.001*c | |||
| Sero -positive | 4 (1.8) | 5 (7.4) | 9 (8.7) | ||
| Sero- negative | 135 (61.4) | 35 (51.5) | 72 (69.2) | ||
| Unknown | 81 (36.8) | 28 (41.2) | 23 (22.1) | ||
| Lung cavitations | 0.85 | 0.547 | |||
| Yes | 70 (31.8) | 24 (35.3) | 38 (36.5) | ||
| No | 118 (53.6) | 34 (50.0) | 49 (47.1) | ||
| Unknown | 32 (14.6) | 10 (14.7) | 17 (16.4) | ||
| Sputum smear | 0.79 | 0.043d | |||
| Positive | 81 (36.8) | 27 (39.7) | 48 (46.2) | ||
| Negative | 110 (50.0) | 34 (50.0) | 51 (49.0) | ||
| Unknown | 29 (13.2) | 7 (10.3) | 5 (4.8) | ||
P-values for chi-square test comparing distribution of characteristics between small and medium cluster groups,
P-values for chi-square test comparing distribution of characteristics between small and large cluster groups.
P-value for chi-square test comparing frequency distribution of HIV status between small and medium cluster groups without unknowns is 0.02.
P-value for chi-square rest comparing frequency distribution of excessive alcohol use between small and large cluster group without unknowns is 0.001.
P-value for variable HIV status between small and large cluster groups after excluding unknowns is 0.01.
P-values for Fisher’s exact test
Table 4.
Multivariate logistic regression model determining risk factors for being in medium clusters (n=68) and large clusters (n=104) with small cluster group (n=220) as the control group.
| Variables | Medium cluster
|
P | Large cluster
|
P | ||
|---|---|---|---|---|---|---|
| OR | 95% CI | OR | 95% CI | |||
| Age | 0.37 | 0.515 | ||||
| <65 years | 1.00 | referent | 1.00 | referent | ||
| ≥ 65 years | 1.57 | 0.59–4.13 | 0.7 | 0.24–2.05 | ||
| Sex | 0.89 | 0.421 | ||||
| Male | 1.00 | referent | 1.00 | referent | ||
| Female | 1.05 | 0.50–2.22 | 0.76 | 0.39–1.49 | ||
| Race | 0.74 | 0.047 | ||||
| Other | 1.00 | referent | 1.00 | referent | ||
| Non-Hispanic black | 1.13 | 0.54–2.36 | 1.89 | 1.01–3.55 | ||
| HIV status | 0.02 | 0.066 | ||||
| seronegative | 1.00 | referent | 1.00 | referent | ||
| seropositive | 5.03 | 1.25–20.14 | 3.31 | 0.93–11.85 | ||
| Homeless within past year | 0.005 | |||||
| No | N/A* | 1.00 | referent | |||
| Yes | 10.35 | 2.02–52.93 | ||||
| Excessive alcohol use within past year | N/A* | 0.486 | ||||
| No | 1.00 | referent | ||||
| Yes | 1.28 | 0.64–2.55 | ||||
Analysis was not applicable because the variables “homelessness within past year” and “excessive alcohol use” were not significantly differently distributed between the medium cluster group and the small cluster group.
Genomic comparison of a large cluster isolate and a non-clustered isolate
To investigate the potential microbial factors involved in clustering, a microarray-based comparative genomic hybridization was performed to identify LSPs in the genomes of an isolate belonging to a large cluster (SA201) and a non-clustered isolate (SA178). The microarray hybridization followed by PCR and DNA sequencing, to determine the exact location of the deletions or sequence variations, identified 12 different LSPs in the genomes of SA201 and SA178 (Table 3). These 12 LSPs ranged in size from 21 bp to 17,793 bp and included six deletions, three IS6110 insertion-mediated deletion events, one repeat of an adjacent region, one gene replacement event, and one variation in a portion of the gene sequence. Four of the twelve LSPs were exactly the same in the two isolates. These four LSPs affected 16 genes encoding two PPE, a PE_PGRS, four transposases, a lipoprotein, a probable conserved pro-, gly-, val-rich secreted protein, a serine-esterase, and six hypothetical proteins. There were three and five LSPs unique to SA201 and SA178, respectively (Table 4). However, two of these unique LSPs (one in SA201 and one in SA178) involved the same genomic region but the deletion was not exactly the same in the two isolates (Table 3).
DISCUSSION
Although the incidence of recently transmitted TB as well as reactivation disease has been decreasing in Arkansas, the major fraction of the decrease has been due to a decrease in the incidence of reactivated infections [7]. These findings highlight the need to know more about the factors involved in active transmission. Host risk factors for clustering identified in this study included non-Hispanic black race and age < 65 years. Furthermore, a significantly higher proportion of large cluster cases were non-Hispanic black, homeless, less than 65 years old, male sex, smear-positive sputum, excessive use of alcohol, smear-positive, and HIV sero-positive, compared to cases in small clusters all diagnosed within one year. However, being non-Hispanic black and homeless within the past year were the only two host characteristics that were identified as independent risk factors for being in large clusters, representing current transmission. Comparing the genomic content of one of the large cluster strains to that of a non-clustered strain from the same community identified 25 genes that differed between the two strains, potentially contributing to the observed differences in transmission.
Two characteristics are identified as risk factors for clustering of M. tuberculosis genotypes in our study, both of which, the non-Hispanic black race and younger age are among the previously known risk factors [8–11]. It is interesting to observe, in the current study, the differences in host risk factors when comparing clustered cases to non-clustered cases and large cluster cases to small cluster cases. Homelessness, a commonly known risk factor for clustering, was not significantly associated with small clusters; however, it was significantly associated with large clusters. It was found previously that M. tuberculosis genotype clusters in our study population represent both clusters resulted from current ongoing tuberculosis transmission, such as the large clusters, and clusters resulted from the reactivation of clusters of cases involved in remote tuberculosis transmission, which are more likely to be seen as small clusters [7, 14]. Our finding suggest that while being non-Hispanic black is a risk factor for both tuberculosis transmission and reactivation, being homeless mainly affect the chance for tuberculosis transmission. Our observation that social/demographic factors (e.g. being non-Hispanic black and homeless) are associated with large clusters, but clinical characteristics (sputum smear positivity, pulmonary cavitary disease, and HIV sero-positivity) are not, despite the inclusion of some large clusters resulting from ongoing outbreaks that might be caused by highly infectious TB cases in the analysis, suggests that social behavioral factors have a more important role in transmission of tuberculosis than does the infectiousness of the source.
Although previously documented and confirmed in this study that host risk factors can play an important role in TB transmission, the ability of M. tuberculosis to be transmitted from host to host is not well understood. Epidemiologic studies have observed that some strains are more successful in transmission than others [9, 15–17]. A large cluster of TB could be explained if the infecting strain has a higher probability of transmission or a higher probability of infection progressing to disease. The mycobacterial cell envelope contains immunomodulatory molecules that are important determinants of intracellular survival and virulence [18]. Two of the genes (MT1800 and MT1802) affected by an LSP in strain SA201, the more widely transmitted strain, but not SA178, the less successful strain encode proteins that influence properties of the cell envelope [19]. MT1802 encodes a membrane protein of the MmpL family. Although the function of this member of the MmpL family has not been studied, these membrane proteins are thought to have a function in the transport of lipids across the cell membrane, affecting the structure of the cell envelope [20]. Six of the 16 genes absent in strain SA201, but present in SA178, were of the PE/PPE gene family. PE/PPE family genes are thought to play a role in maintenance of the latent state through antigenic variation [21]. Disruption of these genes may decrease the available repertoire of antigens available to the M. tuberculosis strain, decreasing its ability to remain in a latent state. The other important LSP found in this study was the presence of one of the M. tuberculosis lipase-encoding genes, lipR (Rv3084) in strain SA201 and absent in strain SA178.
M. tuberculosis lipases comprise a diverse class of enzymes that are involved in lipid metabolism and may, therefore, have an important role in tuberculosis pathogenesis. Recently, as a follow-up to our microarray findings, Sheline and coworkers explored the association of LSP in lipR with patient characteristics using a population-based sample of 665 clinical isolates and found that DNA fingerprinting-clustered cases infected with a lipR LSP isolate were more often epidemiologically linked than clustered cases infected with a lipR wild-type isolate [22]. This finding suggests the usefulness of the genomic comparison conducted in the present study. Further studies are needed to investigate whether the presence or absence of any of these 25 genes is associated with large clusters of tuberculosis cases. This will require a larger number of large cluster strains than is present in ourM. tuberculosis collection. The 25 genes that differ between the large cluster strain SA201 and the non-clustered strain SA178 identified in this study can serve as a basis for additional functional studies or population-based molecular epidemiologic studies that examine the association of these genetic changes with the ability of M. tuberculosis to cause persistent clusters of disease.
Table 5.
Twelve large sequence polymorphisms (LSPs) identified by microarray-based genomic comparison of strain SA201 (cluster pathogen) and strain SA178 (non-cluster pathogen)
| Isolate | Gene(s) having LSP | Positions | Comparis on strain | Size of LSP |
|---|---|---|---|---|
| SA201 | ||||
| MT1799–1814.2 | IS6110 insertion; 1978471–1996263 deleted | CDC1551 | 17,793 bp | |
| MT1836–1839 | deletion begins in MT1836 and ends in MT1839 | CDC1551 | 3,206 bp | |
| MT3098–3101 | 3371306–3374005 replaced with 3375101–3376445 (MT3106) | CDC1551 | 2,700 bp | |
| SA178 | ||||
| MT1803–1812 | IS6110 insertion; 1985524–1993916 deleted | CDC1551 | 8,393 bp | |
| MT0676 | 744075–744607 deleted (same as H37Rv) | CDC1551 | 533 bp | |
| MT2080–2082 | 2266058–2271058 deleted (same as H37Rv) | CDC1551 | 5,001 bp | |
| Rv3083–3085 | 3448497–3451398 deleted | H37Rv | 2,902 bp | |
| MT3429 | IS6110 insertion; 3708984–3709088 deleted | CDC1551 | 105 bp | |
| SA201 & SA178 | ||||
| Rv1435c | 1612642–1612662 repeated (same as CDC1551) | H37Rv | 21 bp | |
| MT2144 | sequence varies for 2341593–2341842 (same as H37Rv) | CDC1551 | 250 bp | |
| MT2619 | 2862617–2863272 deleted (same as H37Rv) | CDC1551 | 656 bp | |
| Rv3425–3428c | Rv3425 – Rv3428c deleted (same as CDC1551) | H37Rv | 4,927 bp | |
Table 6.
Genes with identified differences between SA201 (cluster pathogen) and SA178 (non-cluster pathogen).
| Isolate | Gene having LSP | Product |
|---|---|---|
| SA201 | MT1799–1814.2 | |
| MT1799 | Phospholipase C (plcD) | |
| MT1800 | Glycosyl transferase | |
| MT1801 | Molybdopterin oxidoreductase | |
| MT1802 | Membrane protein, MmpL family | |
| MT1813 | Hypothetical protein | |
| MT1814 | Hypothetical protein | |
| MT1814.2 | Hypothetical protein | |
| MT1836–1839 | ||
| MT1836 | PPE | |
| MT1837 | PE | |
| MT1838 | PPE | |
| MT1838.1 | Hypothetical protein | |
| MT1839 | PPE | |
| MT3098–3101 | ||
| MT3098 | PPE | |
| MT3099 | IS6110 | |
| MT3100 | IS6110 | |
| MT3101 | PPE | |
| SA178 | MT0676 | Alpha-mannosidase |
| MT2080–2082 | ||
| MT2080 | Hypothetical protein | |
| MT2080.1 | Hypothetical protein | |
| MT2081 | Hypothetical protein | |
| MT2082 | Putative helicase | |
| Rv3083–3085 | ||
| Rv3083 | Hypothetical protein | |
| Rv3084 | Lipase (lipR) | |
| Rv3085 | Hypothetical protein | |
| MT3429 | Hypothetical protein |
Acknowledgments
This study was supported by the National Institutes of Health grant NIH-R01-AI151975 and the interagency agreement 98FED10318 between the Veterans Administration, the Centers for Disease Control and Prevention and the Arkansas Department of health. The authors thank all of the colleagues in Dr. Zhenhua Yang’s laboratory for helpful discussions during the laboratory investigation and acknowledge Larissa Andersen’s assistance in verifying cluster assignment.
Footnotes
Conflict of interest: None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.CDC. Decrease in reported tuberculosis cases - United States, 2009. MMWR Morb Mortal Wkly Rep. 2010;59(10):289–294. [PubMed] [Google Scholar]
- 2.CDC. Reported tuberculosis in the United States, 2005. Atlanta, GA: U. S. Department of Health and Human Services, CDC; 2006. [Google Scholar]
- 3.CDC. Reported tuberculosis in the United States. http://www.cdc.gov/nchstp/tb/surv/surv2004/default.htm.
- 4.CDC. Trends in tuberculosis morbidity --- United States, 1992–2002. MMWR. 2003;52(11):217–222. [PubMed] [Google Scholar]
- 5.CDC. Reported Tuberculosis in the United States, 2001. Atlanta: U.S. Department of Health and Human Services; 2002. [Google Scholar]
- 6.CDC. Expanded tuberculosis surveillance and tuberculosis morbidity-United States, 1993. MMWR. 1994;43:361–366. [PubMed] [Google Scholar]
- 7.France AM, Cave MD, Bates JH, Foxman B, Chu T, Yang Z. What’s driving the decline in tuberculosis in Arkansas? A molecular epidemiologic analysis of tuberculosis trends in a rural, low-incidence population, 1997 2003. Am J Epidemiol. 2007;166(6):662–671. doi: 10.1093/aje/kwm135. [DOI] [PubMed] [Google Scholar]
- 8.Small PM, Hopewell PC, Singh SP, Paz A, Parsonnet J, Ruston DC, Schecter GF, Daley CL, Schoolnik GK. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N Engl J Med. 1994;330(24):1703–1709. doi: 10.1056/NEJM199406163302402. [DOI] [PubMed] [Google Scholar]
- 9.Barnes PF, el-Hajj H, Preston-Martin S, Cave MD, Jones BE, Otaya M, Pogoda J, Eisenach KD. Transmission of tuberculosis among the urban homeless. Jama. 1996;275(4):305–307. [PubMed] [Google Scholar]
- 10.Ellis BA, Crawford JT, Braden CR, McNabb SJ, Moore M, Kammerer S. Molecular epidemiology of tuberculosis in a sentinel surveillance population. Emerg Infect Dis. 2002;8(11):1197–1209. doi: 10.3201/eid0811.020403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McNabb SJ, Braden CR, Navin TR. DNA fngerprinting of Mycobacterium tuberculosis: lessons learned and implications for the future. Emerg Infect Dis. 2002;8(11):1314–1319. doi: 10.3201/eid0811.020402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kik SV, Verver S, van Soolingen D, de Haas PE, Cobelens FG, Kremer K, van Deutekom H, Borgdorff MW. Tuberculosis outbreaks predicted by characteristics of first patients in a DNA fingerprint cluster. Am J Respir Crit Care Med. 2008;178(1):96–104. doi: 10.1164/rccm.200708-1256OC. [DOI] [PubMed] [Google Scholar]
- 13.Barnes PF, Cave MD. Molecular epidemiology of tuberculosis. N Engl J Med. 2003;349(12):1149–1156. doi: 10.1056/NEJMra021964. [DOI] [PubMed] [Google Scholar]
- 14.Braden CR, Templeton GL, Cave MD, Valway S, Onorato IM, Castro KG, Moers D, Yang ZH, Stead WW, Bates JH. Interpretation of restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolates from a state with a large rural population. Journal of Infectious Diseases. 1997;175(6):1446–1452. doi: 10.1086/516478. [DOI] [PubMed] [Google Scholar]
- 15.Yang Z, Barnes PF, Chaves F, Eisenach KD, Weis SE, Bates JH, Cave MD. Diversity of DNA fingerprints of Mycobacterium tuberculosis isolates in the United States. J Clin Microbiol. 1998;36(4):1003–1007. doi: 10.1128/jcm.36.4.1003-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dillaha J, Yang Z, Ijaz K, Eisenach K, Cave M, Wilson F, Stead W, Bates J. Transmission over a 54-year time span of an endemic strain of Mycobacterium tuberculosis in hospital, nursing home, jail and community settings. Presented at ATS International Conference; May 5–10, 2000: 2000; Toronto, Canada. 2000. [Google Scholar]
- 17.Ijaz K, Yang Z, Templeton G, Stead WW, Bates JH, Cave MD. Persistence of a strain of Mycobacterium tuberculosis in a prison system. Int J Tuberc Lung Dis. 2004;8(8):994–1000. [PubMed] [Google Scholar]
- 18.Karakousis PC, Bishai WR, Dorman SE. Mycobacterium tuberculosis cell envelope lipids and the host immune response. Cell Microbiol. 2004;6(2):105–116. doi: 10.1046/j.1462-5822.2003.00351.x. [DOI] [PubMed] [Google Scholar]
- 19.Fleischmann RD, Alland D, Eisen JA, Carpenter L, White O, Peterson J, DeBoy R, Dodson R, Gwinn M, Haft D, et al. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J Bacteriol. 2002;184(19):5479–5490. doi: 10.1128/JB.184.19.5479-5490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domenech P, Reed MB, Barry CE., 3rd Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect Immun. 2005;73(6):3492–3501. doi: 10.1128/IAI.73.6.3492-3501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dheenadhayalan V, Delogu G, Sanguinetti M, Fadda G, Brennan MJ. Variable expression patterns of Mycobacterium tuberculosis PE_PGRS genes: evidence that PE_PGRS16 and PE_PGRS26 are inversely regulated in vivo. J Bacteriol. 2006;188(10):3721–3725. doi: 10.1128/JB.188.10.3721-3725.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheline KD, France AM, Talarico S, Foxman B, Zhang L, Marrs CF, Bates JH, Cave MD, Yang Z. Does the lipR gene of tubercle bacilli have a role in tuberculosis transmission and pathogenesis? Tuberculosis (Edinb) 2008 doi: 10.1016/j.tube.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]