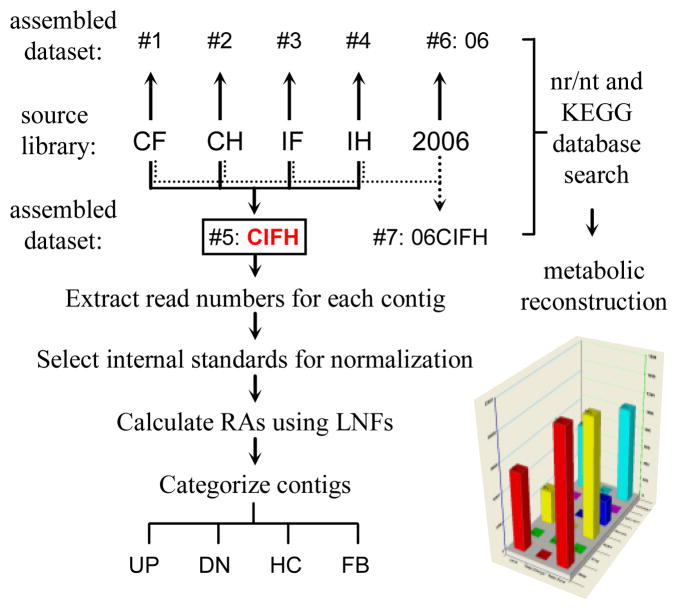
Fig 1. Scheme of library sequencing, dataset assembling, read normalization, contig categorization, and function prediction.
Five cDNA libraries (CF, CH, IF, IH, and 2006) were assembled into seven datasets, one of which (#5: CIFH) was further analyzed by extracting numbers of CF, CH, IF and IH reads assembled into each contig. As described in Section 2.3, read numbers were calibrated using library normalization factors (LNFs) for the calculation of relative abundances (RAs) or adjusted read numbers (ARNs). Based on thresholds set arbitrarily, contigs were categorized into four groups: UP and DN for up- and down-regulated; HC and FB for hemocyte- or fat body-specific.