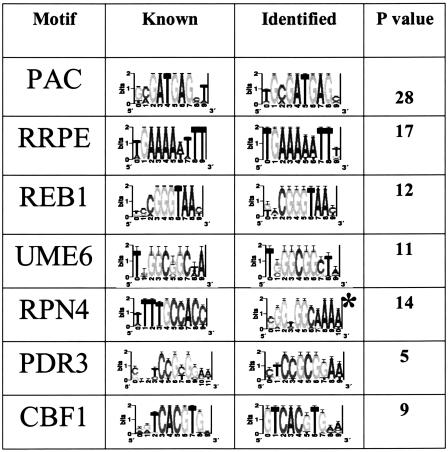
Table 1.
Identification of Known Binding Sites
Columns: (1) binding site (motif) name, (2) sequence-logo representation of known binding site, (3) sequence-logo representation of the best phylogenetically mapped motif (* reverse complement), (4) P-value for network-level conservation (Log10(p)) of the best matching phylogenetically mapped motif.