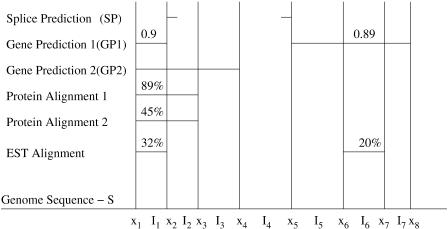
Figure 2.
Partitioned output from three evidence types: splice predictions, gene predictions, and sequence alignments. The five sources of evidence (listed in order from top to bottom) are output from a splice prediction program (SP); a gene prediction program (GP1) with exon confidence scores 0.9 and 0.89; a gene prediction program (GP2) with no confidence scores; 89% and 45% identity alignments from a protein database, which make up a single evidence source; and 32% and 20% identity alignments from an EST database. The genome sequence is divided into intervals defined by each potential boundary x1,x2,...,x8. The non-overlapping intervals I1,...,I7 are used to score gene models. The predicted splice site at x5 is associated with I5.