Fig. X.6.
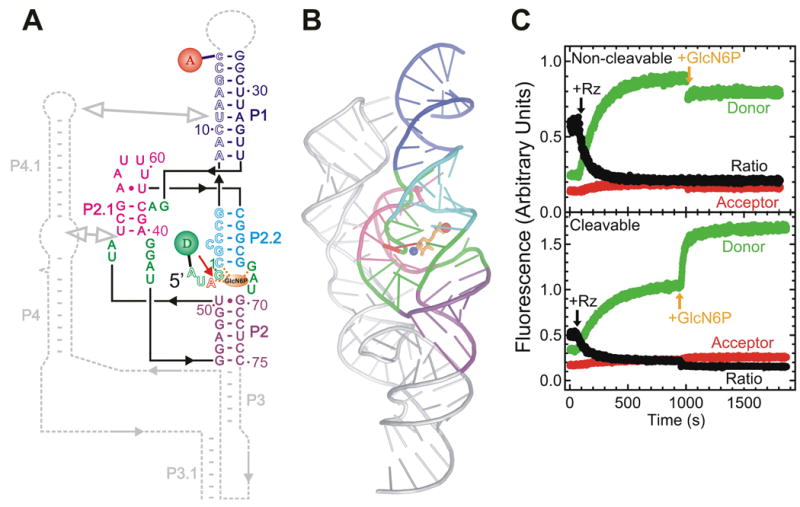
FRET studies of the glmS ribozyme. (A) Secondary structure of the trans-acting glmS ribozyme utilized in these studies. Helices are color coded, the catalytic core is highlighted in green, the cleavage site in the substrate (outlined, S) is marked by a closed red arrow, and the cofactor GlcN6P is shown as an orange oval interacting with several core residues (dashed orange lines). The substrate strand is labeled with donor (D) and acceptor (A) fluorophores, as indicated, which undergo FRET. Dashed gray lines indicate connections and a downstream pseudoknot that are present in the naturally occurring, cis-acting ribozyme. (B) Cartoon representation of the crystal structure of the Thermoanaerobacter tengcongensis glmS ribozyme (PDB ID: 2H0Z) (Klein and Ferre-D'Amare 2006), color-coded as in panel A with removed parts shown in silver. Red sphere, ligand-chelating Mg2+ ion; blue sphere, amino group of the ligand presumably involved in catalysis. (C) Changes over time in donor, acceptor, and acceptor:donor ratio signals upon addition of a 10-fold excess of glmS ribozyme (Rz) and subsequently saturating (10 mM) GlcN6P ligand to either the non-cleavable or cleavable substrate under standard conditions (50 mM HEPES-KOH, pH 7.5, 200 mM K+, 10 mM Mg2+, 25 mM DTT, 25°C), as indicated. In part modified with permission from (Tinsley et al. 2007).
