Abstract
The life of a plant is characterized by major phase transitions. This includes the agriculturally important transitions from seed to seedling (germination) and from vegetative to generative growth (flowering induction). In many plant species, including Arabidopsis thaliana, freshly harvested seeds are dormant and incapable of germinating. Germination can occur after the release of dormancy and the occurrence of favourable environmental conditions. Although the hormonal control of seed dormancy is well studied, the molecular mechanisms underlying the induction and release of dormancy are not yet understood.
In this study, we report the cloning and characterization of the mutant reduced dormancy 2-1 (rdo2-1). We found that RDO2 is allelic to the recently identified dormancy gene TFIIS, which is a transcription elongation factor. HUB1, which was previously called RDO4, was identified in the same mutagenesis screen for reduced dormancy as rdo2-1 and was also shown to be involved in transcription elongation. The human homologues of RDO2 and HUB1 interact with the RNA Polymerase II Associated Factor 1 Complex (PAF1C). Therefore, we investigated the effect of other Arabidopsis PAF1C related factors; VIP4, VIP5, ELF7, ELF8 and ATXR7 on seed dormancy. Mutations in these genes resulted in reduced dormancy, similar to hub1-2 and rdo2-1. Consistent with a role at the end of seed maturation, we found that HUB1, RDO2 and VIP5 are upregulated during this developmental phase. Since mutants in PAF1C related factors are also described to be early flowering, we conclude that these components are involved in the regulation of both major developmental transitions in the plant.
Introduction
Germination and induction of flowering are important developmental switches in the life cycle of plants. Seed dormancy is defined as the incapacity of a viable seed to germinate and evolved in plants to survive periods of unfavourable environmental conditions like dry summers. In many plant species, including the model plant Arabidopsis thaliana, primary seed dormancy is induced during the seed maturation phase and is highest in freshly harvested seeds. Dormancy is released by imbibition of seeds at low temperatures (stratification) or by dry storage (after-ripening). Germination requires the protrusion of the radicle through the surrounding structures (endosperm and testa in Arabidopsis) and can occur when non-dormant seeds meet permissive environmental conditions regarding humidity, light and temperature [1]. The depth of seed dormancy varies within and between plant species. Most important agricultural crop plants show shallow seed dormancy because this has been selected for during the domestication process. In some crops, including cereals, very low dormancy levels can lead to pre-harvest sprouting and consequently reduced product quality [2].
The plant hormone abscisic acid (ABA) is required for the induction of dormancy, whereas germination needs gibberellins (GA). Mutants that affect bioactive levels, or interfere with the signalling pathways of these hormones, usually show seed dormancy phenotypes [3], [4]. Several other hormones also influence dormancy and germination usually by interaction with ABA. Ethylene for instance acts antagonistically to ABA and promotes endosperm rupture [5]. Recently, a role for 12-oxo-phytodienoic acid (OPDA) in germination repression has been identified that is synergistic with ABA [6].
Despite the knowledge at the hormone level, the control of seed dormancy at the molecular level is still poorly understood. To obtain more insight in the molecular processes controlling dormancy, various mutagenesis screens and Quantitative Trait Locus (QTL) analyses have been performed. A major dormancy gene identified both by QTL analysis and mutagenesis screens is DELAY OF GERMINATION 1 (DOG1). DOG1 encodes a protein with unknown function [7]. Another mutagenesis screen yielded four reduced dormancy (rdo) mutants in the Landsberg erecta genetic background [8], [9]. These mutants all have wild-type ABA levels and sensitivity and show only mild pleiotropic effects in the adult plant stage [9]. One of the underlying genes, RDO4, was cloned and renamed HISTONE MONOUBIQUITINATION 1 (HUB1) [10]. HUB1 encodes a C3HC4 RING finger protein, which is required for monoubiquitination of histone H2B. H2B ubiquitination regulates initiation and early elongation steps in transcription, whereas histone H2B deubiquitination is important for transcription elongation. It has been suggested that there might be multiple rounds of ubiquitination and deubiquitination during transcription elongation [11], [12]. Consistent with a role of histone ubiquitination in gene transcription efficiency, absence of functional HUB1 leads to altered expression of several dormancy-related genes [10].
Several other factors are known to play a role in transcription elongation, including Transcription factor S-II (TFIIS). TFIIS is able to overcome transcription arrest by RNA polymerase II and has recently been shown to control seed dormancy in Arabidopsis [13], [14]. In this work we demonstrate that the rdo2 mutation, isolated in the same screen as rdo4/hub1 [9], is allelic to TFIIS. This suggests that HUB1 and RDO2 both influence transcription efficiency. In agreement, a significant overlap in differentially expressed genes during seed maturation was found between both mutants. This confirms their involvement in the same process. Consistent with a role at the end of seed maturation, both genes are upregulated during this phase. The human homologues of RDO2 and HUB1 interact with the RNA Polymerase II Associated Factor 1 Complex (PAF1C) [14]. We show that mutants in several other PAF1C associated genes also have reduced dormancy. These mutants were originally isolated based on their early flowering phenotype. This indicates that PAF1C associated genes are involved in the regulation of both major developmental transitions in the plant.
Results
RDO2 encodes a TFIIS transcription elongation factor
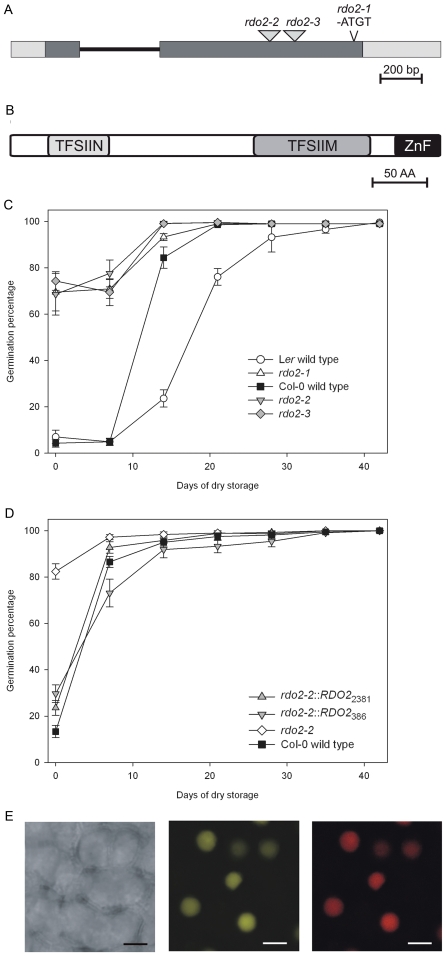
We aimed to identify the rdo2 mutation, which causes reduced dormancy and maps at the bottom of chromosome 2 [9]. To reduce the influence of natural variation between different accessions and to ease the recognition of the mutant phenotype during the mapping process, rdo2-1 was crossed with the Near Isogenic Line (NIL) LCN2-18 [15]. This NIL has a Ler isogenic genetic background, except for a 4.5 Mb introgression of Cvi at the bottom of chromosome 2 containing the RDO2 locus. Using a mapping population of 1100 F2 plants, the location of rdo2-1 could be assigned to a region of 46 kb between the markers T6A23-1 and T6A23-2 located at respectively 16.123 and 16.169 Mb. This region contains 15 annotated genes. Based on the structure of these genes (analyzed in The Arabidopsis Information Resource [16]) and their expression pattern (analyzed with Genevestigator [17]), the candidate gene At2g38560 was selected. Sequencing revealed a four bp deletion at the end of the coding sequence of At2g38560 in the rdo2-1 mutant (Figure 1A). The protein encoded by this gene contains three structural domains, named Transcription factor IIS N-terminal (TFSIIN), Transcription elongation factor S-IIM (TFSIIM) and Zinc finger (ZnF) (Figure 1B). This combination of domains is characteristic for Transcription elongation factor SII (TFIIS) [18]. Due to the 4 bp deletion, the rdo2-1 mutant gene translates into a protein lacking the ZnF domain, which most likely renders it not functional. At2g38560 has previously been identified as a dormancy gene by Grasser and colleagues [13], who named the gene TFIIS.
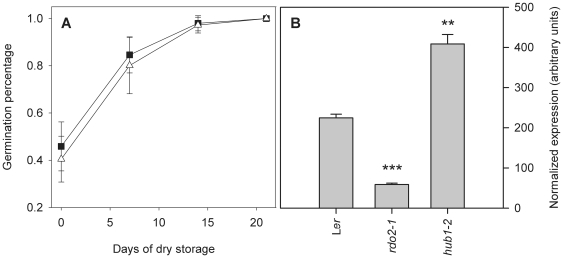
Figure 1. Characterisation of RDO2.
(A) Schematic representation of the RDO2 gene, indicating the positions of the rdo2-1 4 bp deletion and the rdo2-2 and rdo2-3 T-DNA insertions. Exons in the RDO2 locus are represented as grey boxes, UTR regions as white boxes and the intron as a black line. (B) Schematic representation of RDO2 structural protein domains; Transcription factor IIS, N-terminal (TFSIIN), Transcription elongation factor S-IIM (TFSIIM) and Zinc finger (ZnF), obtained by At2g38560 protein analysis with the Simple Modular Architecture Research Tool (SMART) [49]. (C) Dormancy/germination behaviour of rdo2-1 (white triangles up) and wild-type Ler (white circles), rdo2-2 (SALK_027259; grey triangles down), rdo2-3 (SALK_133631; grey diamond) and wild-type Col (black squares). Germination is expressed as percentage of germinated seeds after different periods of seed dry storage starting from harvest. Error bars represent SE, n≥14. (D) Dormancy/germination behaviour of complemented rdo2-2 mutants. The rdo2-2 mutant (SALK_027259; white circles) was complemented with genomic-DNA fragments containing the complete RDO2 coding sequence and 386 bp (rdo2-2::RDO2386; grey triangles down) or 2381 bp (rdo2-2::RDO22381; grey triangles up) upstream of the RDO2 start codon. Col wild-type is shown as black squares. Germination is expressed as percentage of germinated seeds after different times of seed dry storage starting from harvest. Error bars represent SE, n≥14. (E) YFP signal in nuclei of rdo2 mutant plants that were stably transformed with a p2X35S:RDO2:YFP construct. Left panel transmission, middle panel YFP fluorescence, right panel Propidium Iodide staining. Scale bar represents 3 µm.
The identity of At2g38560 as RDO2 was confirmed with additional independent T-DNA insertion mutant alleles (rdo2-2/tfIIs-2 [13] and rdo2-3; Figure 1A) in the Columbia (Col) background. Both insertion mutants lack full-length RDO2 mRNA and showed reduced dormancy, similar to rdo2-1 (Figure 1C). In addition, complementation of rdo2-2 with the RDO2 genomic locus complemented the mutant phenotype (Figure 1D).
RDO2 is ubiquitously expressed throughout all plant tissues as shown in the Arabidopsis eFP browser [19] and [13]. These authors also showed that the Arabidopsis TFIIS protein is localized in the nucleus of transiently transformed protoplasts, which is consistent with a role for RDO2 in transcription elongation [13]. In agreement, we detected YFP signal in nuclei of rdo2-1 mutant plants that were stably transformed with a p2X35S:RDO2:YFP construct (Figure 1E). However, this construct did not complement the rdo2-1 phenotype. This indicates that the YFP tag probably interferes with RDO2 function.
Mutations in HUB1 and RDO2 affect the expression of an overlapping set of genes
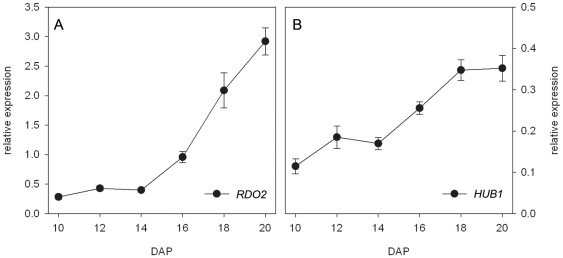
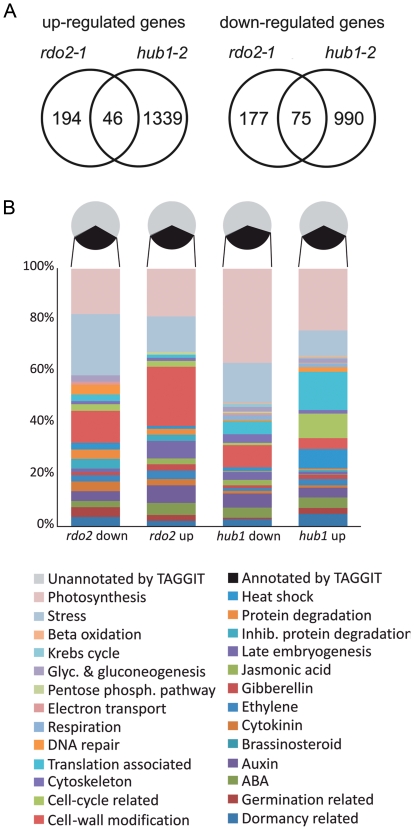
The hub1-2 mutant (previously named rdo4) was identified in the same mutagenesis screen as rdo2-1 [9]. HUB1 is required for monoubiquitination of histone H2B [10]. This histone modification is involved in transcription initiation and elongation [12], which suggests that RDO2 and HUB1 are involved in the same process. The RDO2 and HUB1 genes are both ubiquitously expressed. Because their mutants show reduced seed dormancy, we analyzed their expression dynamics in detail during seed maturation. RT-PCR analysis indicated that both genes are strongly upregulated during this phase (Figure 2A, B). This increase in expression levels, together with the identity of HUB1 and RDO2 as transcription initiation and elongation factors and the observation that rdo2-1 and hub1-2 mutants have reduced dormancy levels, indicates that transcription maintenance towards the end of seed maturation is probably required for the induction of seed dormancy. Therefore, we analyzed the transcriptomes of nearly ripe siliques (18–19 DAP) of the hub1-2 and rdo2-1 mutants in comparison with wild-type Ler using Affymetrix GeneChip Arabidopsis ATH1 Genome Micro-Arrays. The hub1-2 and rdo2-1 mutants revealed respectively 2450 and 492 differentially expressed genes (Benjamini & Hochberg (BH) adjusted P-value<0.01) (Dataset S1). The hub1-2 mutant thus has a stronger influence on the transcriptome than rdo2-1. A relatively high number of differentially expressed genes (46 up- and 75 downregulated) overlapped between both mutants (Figure 3A). The significance of this overlap was determined by calculating the representation factor, which is the number of overlapping genes divided by the expected number of overlapping genes drawn from two independent random picked groups [20]. The representation factor for upregulated genes in rdo2-1 and hub1-2 is 3.1 (p<3.243e-12) and for downregulated genes 6.4 (p<1.268e-39). HUB1 and RDO2 are both positive regulators of transcription and direct targets of these proteins are expected to be found among the downregulated genes. The downregulated genes indeed showed the highest overlap between hub1-2 and rdo2-1 (30% of the total number of downregulated genes in rdo2-1 overlaps with hub1-2, compared to 19% overlap for upregulated genes). One of the downregulated genes in both hub1-2 and rdo2-1 is the dormancy gene DOG1 (Figure S1). DOG1 protein is required for the induction of dormancy and differences in DOG1 expression can explain differences in dormancy levels [7]. Therefore, downregulation of DOG1 likely contributes to the reduced dormancy of hub1-2 and rdo2-1.
Figure 2. HUB1 and RDO2 transcription during seed maturation.
(A–B) Relative expression of (A) RDO2 and (B) HUB1 in Ler during seed maturation at 10–20 days after pollination (DAP) compared to ACTIN8 expression. Error bars represent SE, n≥3.
Figure 3. Transcriptome analysis of rdo2-1 and hub1-2 siliques at 18–19 DAP.
(A) Venn diagram showing the number of overlapping and unique up- and downregulated genes in rdo2-1 and hub1-2. (B) TAGGIT gene ontology classification of up- and downregulated genes in rdo2-1 and hub1-2.
We used the seed-specific gene ontology classification, called TAGGIT [21], to analyze the differentially regulated genes (Figure 3B). Stress related genes are mainly found among the down-regulated genes of both mutants. The rdo2-1 transcriptome is characterized by an upregulation of cell-wall modifying and late-embryogenesis genes. The hub1-2 transcriptome shows an upregulation of translation associated, cell-cycle related and heat shock genes.
Predicted PAF1C associated factors are upregulated during seed maturation and are required for the induction of seed dormancy
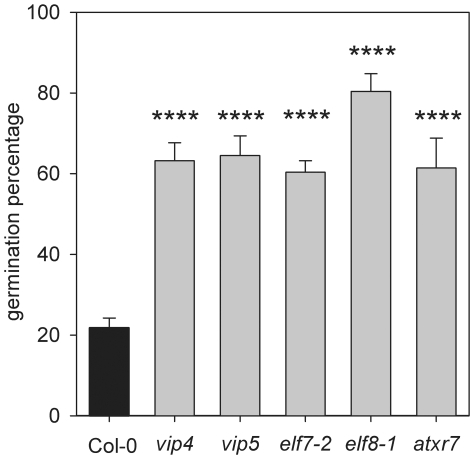
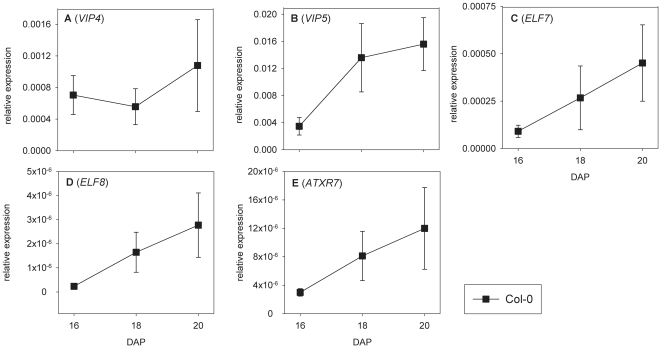
The human homologues of HUB1 and RDO2 are Bre1 and TFIIS respectively. Both interact with the human RNA Polymerase II Associated Factor 1 Complex (PAF1C) [14], [29], which provides a platform for the association of complexes that modulate the structure of chromatin during transcription elongation [30]. Accordingly, PAF1C has a crucial role in the regulation of histone monoubiquitination and is required for recruitment of Set1 and Set2 proteins. These proteins are involved in methylation of histone H3 at respectively K4 and K36, which are activating epigenetic marks for transcription [30]. VERNALIZATION INDEPENDENCE 4 (VIP4), VIP5, EARLY FLOWERING 7 (ELF7) and ELF8 are the Arabidopsis homologues of respectively the yeast proteins Leo1, Rtf1, PAF1 and CTR9, which are all components of PAF1C [31], [32], [33]. ARABIDOPSIS TRITHORAX-RELATED 7 (ATXR7) is the Arabidopsis homologue of Set1 [34]. As shown in Figure 4, the vip4, vip5, elf7-2, elf8-1 and atxr7 mutants all show significantly (p<0.0001) reduced seed dormancy levels, similar to rdo2-1 and hub1-2. Alike HUB1 and RDO2, most of these genes also show a tendency towards upregulation at the end of seed maturation between 16 and 20 DAP (Figure 5), which is however only significant for VIP5 (p = 0.0255) (Figure 5B).
Figure 4. RNAPII associated factors are required to establish dormancy.
Dormancy/germination behaviour of freshly harvested seeds of Col wild-type (black bar), vip4, vip5, elf7-2, elf8-1 and atxr7-1 (gray bars). Error bars represent SE, n≥10, except elf8-1 n = 3; Error bars represent SE. Significance levels: ****p<0.0001; 2-tailed Student's T-test, compared to Col-0.
Figure 5. PAFIC associated factors are upregulated during seed maturation.
Transcript abundance of (A) VIP4, (B) VIP5, (C) ELF7, (D) ELF8, (E) ATXR7, during seed maturation at 16–20 days after pollination (DAP) compared to ACTIN8 expression. Error bars represent SE, n = 4.
VIP4, VIP5, ELF7, ELF8 and ATXR7 are all required for expression of the flowering repressor FLC [28] and their corresponding mutants are early flowering [32], [33], [35]. In addition, FLC expression is decreased in the hub1-4 mutant and hub1 and tfIIs mutants are early flowering [13], [22], [23]. FLC may therefore represent a connection between the regulation of flowering time and seed dormancy as it is expressed in seeds and has been shown to promote germination at low temperatures [36]. However, we could not detect an altered seed dormancy phenotype in the flc mutant (Figure 6A). Moreover, our microarray data indicate that FLC expression in mature siliques is 3-times lower in rdo2-1, but 2-times higher in hub1-2 compared to the wild-type (Figure 6B). Therefore, it is not likely that the reduced dormancy phenotype of the studied mutants is caused by altered FLC expression.
Figure 6. FLC does not affect seed dormancy.
(A) Dormancy/germination behaviour of Col (black squares) and flc101 (open triangles). Germination is expressed as percentage of germinated seeds after different times of seed dry storage. Error bars represent SE, n = 15. (B) FLC expression in siliques, 18–19 days after pollination (DAP) of wild-type Ler and the rdo2-1 and hub1-2 mutants. Expression data were obtained from the microarray experiment described in this study. Significance levels: **p<0.01, ***p<0.001; 2-tailed Student's T-test, compared to wild type Ler.
Discussion
The plant's life cycle is controlled by the timing of two major developmental transitions, germination and induction of flowering. The molecular pathways that control flowering and its interaction with the environment are well studied [37]. However, the control of germination by seed dormancy is still poorly understood at the molecular level. A mutagenesis screen for reduced dormancy in the Ler background yielded four mutants [9], of which the first, hub1-2, was recently cloned [10]. Here, we report the cloning and characterization of a second mutant, rdo2-1. Interestingly, both identified genes are predicted to function in transcription elongation and to associate with the PAF1 complex. Accordingly, transcriptome analysis showed a highly significant overlap in the differentially expressed genes of both mutants. This motivated us to analyse the germination behaviour of additional mutants in genes related to PAF1C, which indeed all showed reduced seed dormancy.
RDO2 encodes a TFIIS transcription elongation factor
The rdo2 mutant was isolated based on its reduced seed dormancy, but also shows some mild additional phenotypes including earlier flowering [8], [13]. The rdo2-1 mutation consists of a 4 bp deletion in a gene encoding a protein with high homology to yeast and human TFIIS (Figure 1B). Originally, TFIIS was isolated as a factor that can stimulate RNA synthesis by specifically stimulating RNAPII [38]. It has been reported that the Arabidopsis TFIIS gene can partially complement the yeast tfIIs mutant [13], indicating that Arabidopsis RDO2 is a bona fide TFIIS transcription elongation factor.
Gene transcription is not only controlled by the recruitment of RNAPII to the promoter, but also by the elongation speed of the moving RNAPII along the coding strand [30]. RNAPII can become paused during elongation at certain sites, which is likely determined by the strength of histone-DNA contacts. This pausing leads to backtracking of the RNAPII, which can cause a complete arrest of transcription [39]. TFIIS helps to overcome such an arrest by stimulating a cryptic, nascent RNA cleavage activity intrinsic to RNAPII [18], [30]. PAF1 is an evolutionary conserved elongation factor complex that was shown to affect transcription elongation efficiency by H2B ubiquitination and H3K4 and H3K79 methylation. It was recently shown that PAF1C also has a more direct role in transcription elongation by its direct interaction with TFIIS and cooperative binding to RNAPII in human HeLa cells [14].
The Arabidopsis TFIIS gene has previously been described as a dormancy gene by Grasser and colleagues [13]. However, in their work increased germination could only be observed after removal of immature seeds at 15 DAP from siliques and freshly harvested seeds germinated at equal rates in the wild-type and tfIIs mutants. In contrast to this, we observed a clear dormancy phenotype in freshly harvested seeds for all three studied rdo2 mutant alleles (Figure 1C). One of these mutants, rdo2-2, is identical to tfIIs-2, for which dormancy phenotype was detected previously in freshly harvested seeds [13]. The different dormancy phenotypes could be explained by differences in the growth conditions because dormancy levels are strongly influenced by the environment.
The influence of RDO2 and HUB1 on gene transcription in maturing seeds
RDO2 and HUB1 are both predicted to influence gene transcription. Our transcriptome analysis showed that indeed relatively high numbers of genes are differentially expressed in the rdo2-1 and hub1-2 mutants (Figure 3A). The number of differentially expressed genes is about five times higher in the hub1-2 mutant compared to the rdo2-1 mutant. This indicates that absence of histone H2B ubiquitination has a stronger impact on gene expression than absence of TFIIS dependent transcription elongation. RDO2 and HUB1 are both positive regulators of transcription and direct target genes of these factors are therefore expected to be down-regulated in the rdo2-1 and hub1-2 mutants. Surprisingly, the number of up-regulated genes is similar to the number of down-regulated genes for both rdo2-1 and hub1-2 (Figure 3A). This indicates that a high number of the differentially expressed genes are probably indirect targets of RDO2 and HUB1. In contrast, a higher number of genes down-regulated was found in the tfIIs mutant compared to the number of up-regulated genes [13]. This difference with our transcriptome analysis could be explained by the different material that was used for the experiments. Grasser and colleagues analysed seedlings for their transcriptome analysis [13], whereas we used siliques at 18–19 DAP. Interestingly, we found an upregulation of the flowering repressor gene FLC in siliques of hub1, whereas earlier studies showed a downregulation of FLC in hub1 seedlings [22], [23]. This underlines that the influence of HUB1 on transcription is different between seeds and seedlings.
We found a relatively high number of down-regulated ‘stress related’ genes in both rdo2-1 and hub1-2 (Figure 3B). Genes belonging to this TAGGIT class are normally upregulated during seed maturation, probably due to the stress conditions caused by desiccation of the maturing seed [24]. These genes could be direct targets of RDO2 and HUB1 and might require these elongation factors to obtain sufficiently high expression levels towards the end of seed maturation. Despite the reduced expression levels of stress related genes in rdo2-1 and hub1-2 mutants, we have not observed any obvious stress related phenotype under our growth conditions. The only clear seed phenotype in rdo2-1 and hub1-2 mutants is reduced dormancy, which can be partially explained by down-regulation of DOG1 in both mutants (Figure S1).
The rdo2-1 transcriptome is characterized by an upregulation of cell-wall modifying and late-embryogenesis genes. The hub1-2 transcriptome shows an upregulation of translation associated, cell-cycle related and heat shock genes (Figure 3B). Genes that regulate cell-wall modification, translation and cell-cycle are upregulated during after-ripening in wild type seeds, probably causing an increased germination potential [25]. Therefore, upregulation of these genes in rdo2-1 and hub1-2 could be contributing to the reduced dormancy of these mutants. In contrast, late-embryogenesis and heat shock genes are associated with dormant expression patterns [25] and their increased expression in rdo2 and hub1 indicates that they are probably independent of the reduced dormancy phenotype.
PAF1C associated factors control both germination and flowering time
RDO2 is a single copy gene in Arabidopsis and highly conserved among eukaryotes. Despite its high conservation and its role in an essential process, the mutant phenotype is weak in Arabidopsis. Weak mutant phenotypes for TFIIS are found in more eukaryotes. S. cerevisiae TFIIS null mutants for instance only show sensitivity to 6-azauracil [26]. In contrast, mice that lack TFIIS die during embryo development at the mid-gestation phase [27]. Similar to rdo2 mutants, the Arabidopsis mutants hub1, elf7-2, elf8-1, vip4, vip5 and atxr7 all show no, or weak, pleiotropic phenotypes [28], [31], [32], [35]. The lack of a strong phenotype for all these mutants is probably due to the presence of multiple elongation factors in eukaryotic cells that function both cooperatively and redundantly [30]. However, this redundancy does not completely compensate for negative effects on transcription of genes required for flowering time and dormancy. Alternatively, these PAF1C associated proteins are only required for transcription of a subset of all genes.
Our data suggest that PAF1C associated factors are required to facilitate expression during late seed maturation, since genes encoding predicted PAF1C associated factors showed a trend towards increased expression during seed maturation (Figures 2 and 6). It is unlikely that increased gene expression at the end of seed maturation is a general phenomenon, as it has been shown that the end of seed maturation is characterized by decreased metabolic activities, including gene transcription [40].
The upregulation of PAF1C associated genes at the end of seed maturation and the clearly reduced dormancy phenotypes of their mutants indicate that they might be especially important in this phase, possibly by counteracting negative effects of desiccation on gene expression.
Conclusion
Overall, our data indicate that PAF1C associated factors are involved in both the control of flowering time and dormancy/germination. They regulate flowering by controlling FLC expression and dormancy by control of the expression of yet unidentified genes. FLC is a flowering repressor that is downregulated by vernalisation and its expression has to be reset every generation. It has been shown that FLC expression is reactivated during embryogenesis [41], [42]. Therefore, the PAF1C associated factors probably control the expression of FLC and dormancy genes simultaneously during seed development. A role for PAF1C associated genes, including the here reported RDO2 gene, as factors regulating both flowering time and seed dormancy could have ecological implications. The moment when a seed germinates will determine the environmental conditions (especially daylength and temperature) to which the plant will be exposed during further growth and thereby indirectly influences life-history traits, including flowering time [43]. Factors like RDO2 and HUB1 could therefore be part of a mechanism that links the germination time to the flowering time, in order to obtain maximum fitness.
Materials and Methods
Plant materials and growth conditions
The rdo2-1 (Ler) mutant is described by [9], hub1-2 (Ler) by [10], vip4 (Col) by [31] and vip5 (Col) by [33]. The elf7-2 and elf8-1 (Col) mutants [32] and LCN2-18 [15] were kind gifts of the authors who described the lines. The rdo2-2/tfIIs-2 [13] (SALK_027259), rdo2-3 (SALK_133631) and atxr7-1 (SALK_14691c) mutants were obtained from the SALK T-DNA insert collection [44]. The rdo2-2 mutant contained a T-DNA insert at 1073 bp, rdo2-3 at 1195 bp and atxr7-1 at 782 bp downstream of the start codon. Full-length mRNAs of the respective genes could not be detected in these mutants. For complementation analysis, the Ler RDO2 genomic locus (including 386 or 2381 bp upstream of the RDO2 start codon) was cloned into the vector pGW-MCS_nos and stably transformed into rdo2-2. Confocal microscopy (Leica TCS SP2, Germany) was used to detect the YFP signal in rdo2-1 mutant plants that were stably transformed with a p2X35S:RDO2:YFP construct generated using the pENSG-YFP [45] vector. Propidium Iodide was used for counterstaining.
Plants were grown on soil containing a mixture of substrate and vermiculite (3∶1). Plants for the germination tests and transcript analyses were grown in Elbanton growth cabinets (Elbanton BV, Kerkdriel, the Netherlands) in long day conditions (16 h light at 22°C and 8 h dark at 16°C). Plants for mapping and crossings were grown in an air-conditioned greenhouse with a day temperature of 20°C and a night temperature of 18°C; 16 h of light was provided daily.
Germination tests
Approximately 50 seeds of individually harvested plants were sown on filter paper, put into transparent moisturized containers and incubated in a germination cabinet (Van den Berg Klimaattechniek, Montfoort, the Netherlands) in long-day conditions (16 h light at 25°C, followed by 8 h darkness at 20°C). After 7 d of incubation, the germination percentages were analyzed with a ‘Germinator’ setup and analyzed as described in [46]. After-ripening conditions of dry seed batches occurred in darkness at 21°C, 50% RH in a controlled cabinet (MMM Medcenter, Brno, Czech Republic).
Transcriptomics
Extraction of RNA from 18–19 DAP siliques was performed using RNAqueous columns (Ambion, Austin, TX, USA). Affymetrix GeneChip Arabidopsis ATH1 Genome Array microarray hybridization and subsequent analysis was performed in house. For all microarray experiments, RNA from three independent biological replicates was used for hybridization and subsequent analyses. Processing and statistical analysis of the microarray data were done in Expressionist Pro v5.1 (Genedata AG, Basel, Switzerland). We used the GC-RMA algorithm for background correction, normalization and probe summarization. Various quality metrics were examined to exclude quality problems. All microarray data are MIAME compliant and have been deposited at the Gene Expression Omnibus database (GEO Accession number: GSE28446).
Control probe sets and probe sets with MAS5 detection P-value smaller than 0.05 in less than two of the nine arrays were filtered out prior to further analysis, leaving 14,655 probe sets. Differential expression of genes (Dataset S1) between each mutant and Ler was assessed using the regularized Bayesian T-test CyberT [47], [48]. BH adjusted P-value (false discovery rate) was adopted for correction of multiple testing [48]. BH adjusted P-value<0.01 was taken as criteria for differential expression.
For the statistical significance of the overlap between two groups of genes we used a web-based tool at http://elegans.uky.edu/MA/progs/overlap_stats.html. We used the seed-specific gene ontology classification program, called TAGGIT [21], to analyze the differentially regulated genes.
Q-RT PCR
RNA from seeds was extracted using RNAqueous small scale Phenol-free total RNA isolation kit in addition with RNA isolation aid (Ambion, Austing, TX, USA). After elution (95°C warm elution buffer), the RNA was cleaned via a high salt precipitation (1.2 M Tri-Na citrate-dihydrate +0.8 M NaCl), washed with 70% ethanol, dried and dissolved. Thereafter, the RNA was precipitated using 25 M LiCl, rinsed with 2 M LiCl, washed with 70% ethanol, dried and dissolved. cDNA synthesis was proceeded with QuantiTect Reverse Transcription Kit (Qiagen, Cat No.205311) including DNAse treatment (gDNA wipeout buffer). Quantitative RT-PCR was subsequently performed via standard procedures using QuantiTect SYBR Green PCR Kit (Qiagen, Cat No.204143) on an Eppendorf Mastercycler realplex2, epgradient cycler. Expression was calculated relative to ACT8 (AT1G49240). All primers used in this study can be found in Table S1. All primers were BLAST searched against the Arabidopsis genome to check uniqueness.
Supporting Information
DOG1 is downregulated in rdo2-1 and hub1-2 . DOG1 expression in siliques, 18–19 days after pollination (DAP) of wild-type Ler and the rdo2-1 and hub1-2 mutants. Expression data were obtained from the microarray experiment described in this study. Significance levels: *p<0.05 **p<0.01; 2-tailed Student's T-test, compared to wild type Ler.
(TIF)
Primer combinations used for RT-PCR analysis.
(DOC)
Differentially expressed genes in the hub1-2 and rdo2-1 mutants.
(XLS)
Acknowledgments
The authors thank Bruno Hüttel for performing the microarray experiments. We thank Yuehui He and Leonie Bentsink for kindly sharing mutant lines, and Anton J.M. Peeters for critical reading of the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Financial support was provided by an EMBO fellowship (ALTF 700-2010) to MvZ (http://www.embo.org/) and by the Deutsche Forschungsgemeinschaft (SO 691/3-1) (http://www.dfg.de/index.jsp) and the Max Planck Society (http://www.mpg.de/de) to WJJS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Finch-Savage WE, Leubner-Metzger G. Seed dormancy and the control of germination. New Phytol. 2006;171:501–523. doi: 10.1111/j.1469-8137.2006.01787.x. [DOI] [PubMed] [Google Scholar]
- 2.Gubler F, Millar AA, Jacobsen JV. Dormancy release, ABA and pre-harvest sprouting. Curr Opin Plant Biol. 2005;8:183–187. doi: 10.1016/j.pbi.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 3.Finkelstein R, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy. Annu Rev Plant Biol. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- 4.Holdsworth MJ, Bentsink L, Soppe WJJ. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008;179:33–54. doi: 10.1111/j.1469-8137.2008.02437.x. [DOI] [PubMed] [Google Scholar]
- 5.Linkies A, Müller K, Morris K, Turečková V, Wenk M, et al. Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: a comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell. 2009;2:3803–3822. doi: 10.1105/tpc.109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dave A, Hernández ML, He Z, Andriotis VME, Vaistij FE, et al. 12-Oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. Plant Cell. 2011;23:583–599. doi: 10.1105/tpc.110.081489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bentsink L, Jowett J, Hanhart CJ, Koornneef M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:17042–17047. doi: 10.1073/pnas.0607877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Léon-Kloosterziel KM, van de Bunt GA, Zeevaart JAD, Koornneef M. Arabidopsis mutants with a reduced seed dormancy. Plant Physiol. 1996;110:233–240. doi: 10.1104/pp.110.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peeters AJM, Blankestijn-de Vries H, Hanhart CJ, Léon-Kloosterziel KM, Zeevaart JAD, et al. Characterization of mutants with reduced seed dormancy at two novel rdo loci and a further characterization of rdo1 and rdo2 in Arabidopsis. Physiol Plant. 2002;115:604–612. doi: 10.1034/j.1399-3054.2002.1150415.x. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Koornneef M, Soppe WJJ. The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell. 2007;19:433–444. doi: 10.1105/tpc.106.049221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pavri R, Zhu B, Li G, Trojer P, Mandal S, et al. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 12.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 13.Grasser M, Kane CM, Merkle T, Melzer M, Emmersen J, et al. Transcript elongation factor TFIIS is involved in Arabidopsis seed dormancy. J Mol Biol. 2009;386:598–611. doi: 10.1016/j.jmb.2008.12.066. [DOI] [PubMed] [Google Scholar]
- 14.Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell. 2010;140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keurentjes JB, Bentsink L, Alonso-Blanco C, Hanhart CJ, Blankestijn-De Vries H, et al. Development of a Near Isogenic Line population of Arabidopsis thaliana and comparison of mapping power with a Recombinant Inbred Line population. Genetics. 2007;175:891–905. doi: 10.1534/genetics.106.066423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Hernandez M, Berardini TZ, Chen G, Crist D, Doyle A, et al. TAIR: a resource for integrated Arabidopsis data. Funct Integr Genomics. 2002;2:239–253. doi: 10.1007/s10142-002-0077-z. [DOI] [PubMed] [Google Scholar]
- 17.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wind M, Reines D. Transcription elongation factor SII. BioEssays. 2000;22:327–336. doi: 10.1002/(SICI)1521-1878(200004)22:4<327::AID-BIES3>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, et al. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim SK, Lund J, Kiraly M, Duke K, Jiang M, et al. A gene expression map for Caenorhabditis elegans. Science. 2001;293:2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- 21.Carrera E, Holman T, Medhurst A, Peer W, Schmuths H, et al. Gene expression profiling reveals defined functions of the ATP-binding cassette transporter COMATOSE late in phase II of germination. Plant Physiol. 2007;143:1669–1679. doi: 10.1104/pp.107.096057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao Y, Dai Y, Cui S, Ma L. Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell. 2008;20:2586–2602. doi: 10.1105/tpc.108.062760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu X, Jiang D, Wang Y, Bachmair A, He Y. Repression of the floral transition via histone H2B monoubiquitination. Plant J. 2009;57:522–533. doi: 10.1111/j.1365-313X.2008.03709.x. [DOI] [PubMed] [Google Scholar]
- 24.Hoekstra FA, Golovina EA, Buitink J. Mechanisms of plant desiccation tolerance. Trends Plant Sci. 2001;6:431–438. doi: 10.1016/s1360-1385(01)02052-0. [DOI] [PubMed] [Google Scholar]
- 25.Carrera E, Holman T, Medhurst A, Dietrich D, Footitt S, et al. Seed after-ripening is a discrete developmental pathway associated with specific gene networks in Arabidopsis. Plant J. 2008;53:214–224. doi: 10.1111/j.1365-313X.2007.03331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakanishi T, Nakano A, Nomura K, Sekimizu K, Natori S. Purification, gene cloning, and gene disruption of the transcription elongation factor S-II in Saccharomyces cerevisiae. J Biol Chem. 1992;267:13200–13204. [PubMed] [Google Scholar]
- 27.Nagata M, Ito T, Arimitsu N, Koyama H, Sekimizu K. Transcription arrest relief by S-II/TFIIS during gene expression in erythroblast differentiation. Genes to Cells. 2009;14:371–380. doi: 10.1111/j.1365-2443.2008.01277.x. [DOI] [PubMed] [Google Scholar]
- 28.Berr A, Xu L, Gao J, Cognat V, Steinmetz A, et al. SET DOMAIN GROUP25 encodes a histone methyltransferase and is involved in FLOWERING LOCUS C activation and repression of flowering. Plant Physiol. 2009;151:1476–1485. doi: 10.1104/pp.109.143941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J, Guermah M, McGinty RK, Lee J-S, Tang Z, et al. RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Bio. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H, van Nocker S. The VERNALIZATION INDEPENDENCE 4 gene encodes a novel regulator of FLOWERING LOCUS C. Plant J. 2002;31:663–673. doi: 10.1046/j.1365-313x.2002.01380.x. [DOI] [PubMed] [Google Scholar]
- 32.He Y, Doyle MR, Amasino RM. PAF1-complex-mediatd histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 2004;18:2774–2784. doi: 10.1101/gad.1244504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh S, Zhang H, Ludwig P, van Nocker S. A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. Plant Cell. 2004;16:2940–2953. doi: 10.1105/tpc.104.026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avramova Z. Evolution and pleiotropy of TRITHORAX function in Arabidopsis. Int J Dev Biol. 2009;53:371–381. doi: 10.1387/ijdb.082664za. [DOI] [PubMed] [Google Scholar]
- 35.Tamada Y, Yun J-Y, Woo SC, Amasino RM. ARABIDOPSIS TRITHORAX-RELATED7 is required for methylation of lysine 4 of histone H3 and for transcriptional activation of FLOWERING LOCUS C. Plant Cell. 2009;21:3257–3269. doi: 10.1105/tpc.109.070060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiang GCK, Barua D, Kramer EM, Amasino RM, Donohue K. Major flowering time gene, FLOWERING LOCUS C, regulates seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2009;106:11661–11666. doi: 10.1073/pnas.0901367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michaels SD. Flowering time regulation produces much fruit. Curr Opin Plant Biol. 2009;12:75–80. doi: 10.1016/j.pbi.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sekimizu K, Kobayashi N, Mizuno D, Natori S. Purification of a factor from Ehrlich Ascites tumor cells specifically stimulating RNA Polymerase II. Biochemistry. 1976;15:5064–5070. doi: 10.1021/bi00668a018. [DOI] [PubMed] [Google Scholar]
- 39.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 40.Comai L, Harada JJ. Transcriptional activities in dry seed nuclei indicate the timing of the transition from embryogeny to germination. Proc Natl Acad Sci USA. 1990;87:2671–2674. doi: 10.1073/pnas.87.7.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi J, Hyun Y, Kang M-J, Yun HI, Yun J-Y, et al. Resetting and regulation of FLOWERING LOCUS C expression during Arabidopsis reproductive development. Plant J. 2009;57:918–931. doi: 10.1111/j.1365-313X.2008.03776.x. [DOI] [PubMed] [Google Scholar]
- 42.Sheldon CC, Hills MJ, Lister C, Dean C, Dennis ES, et al. Resetting of FLOWERING LOCU C expression after epigenetic repression by vernalization. Proc Natl Acad Sci USA. 2008;105:2214–2219. doi: 10.1073/pnas.0711453105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Donohue K. Germination timing influences natural selection on life-history characters in Arabidopsis thaliana. Ecology. 2002;83:1006–1016. [Google Scholar]
- 44.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 45.Jakoby MJ, Weinl C, Pusch S, Kuijt SJH, Merkle T, et al. Analysis of the subcellular localization, function, and proteolytic control of the Arabidopsis cyclin-dependent kinase inhibitor ICK1/KRP1. Plant Physiol. 2006;141:1293–1305. doi: 10.1104/pp.106.081406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joosen RVL, Kodde J, Willems LAJ, Ligterink W, Van Der Plas LHW, et al. GERMINATOR: a software package for high-throughput scoring and curve fitting of Arabidopsis seed germination. Plant J. 2010;62:148–159. doi: 10.1111/j.1365-313X.2009.04116.x. [DOI] [PubMed] [Google Scholar]
- 47.Baldi P, Long AD. A bayesian framework for the analysis of microarray expression data: regularized t-Test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 48.Benjamini Y, Hochberg Y. Controlling the false discovery rate–a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodo. 1995;57:289–300. [Google Scholar]
- 49.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2008;37:229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DOG1 is downregulated in rdo2-1 and hub1-2 . DOG1 expression in siliques, 18–19 days after pollination (DAP) of wild-type Ler and the rdo2-1 and hub1-2 mutants. Expression data were obtained from the microarray experiment described in this study. Significance levels: *p<0.05 **p<0.01; 2-tailed Student's T-test, compared to wild type Ler.
(TIF)
Primer combinations used for RT-PCR analysis.
(DOC)
Differentially expressed genes in the hub1-2 and rdo2-1 mutants.
(XLS)