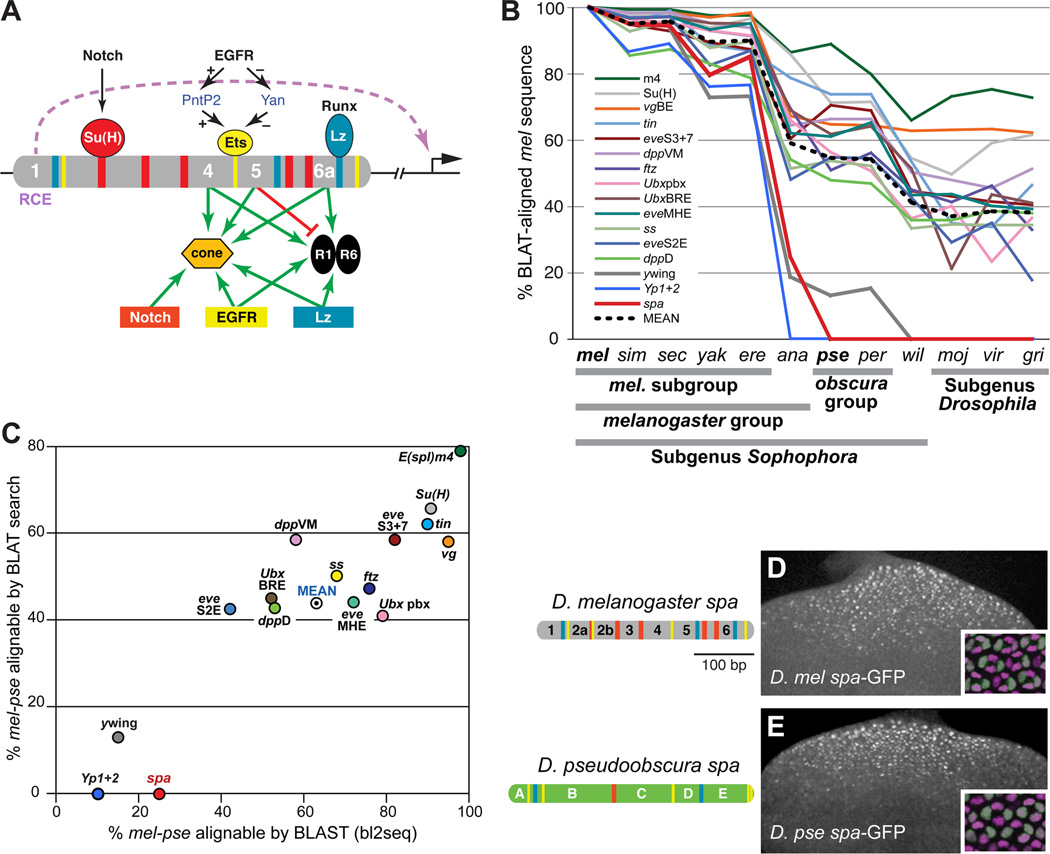
Figure 1. Despite Rapid Sequence Divergence, the Function and Cell-Type Specificity of the sparkling Enhancer Is Conserved.
(A) Summary of the regulatory logic of the sparkling (spa) cone cell enhancer of dPax2. Colored bars indicate known binding sites for Su(H), PntP2/Yan, and Lozenge (Lz); essential novel regulatory sequences 1, 4, 5, and 6a are numbered. RCE indicates the Remote Control Element in region 1 [8].
(B) Comparison of pairwise ortholog sequence similarity between D. melanogaster (mel) and 11 other sequenced Drosophila species, for 16 developmental enhancers. sim, D. simulans; sec, D. sechellia; yak, D. yakuba; ere, D. erecta; ana, D. ananassae; pse, D. pseudoobscura; per, D. persimilis; wil, D. willistoni; moj, D. mojavensis; vir, D. virilis; gri, D. grimshawi. Similarity is measured as the percentage of mel enhancer sequence that is aligned to the orthologous region of the comparison genome by BLAT. See Supplemental Experimental Procedures for detailed information on these reference enhancers.
(C) mel-pse pairwise sequence similarity for 16 developmental cis-regulatory sequences, measured as the percentage of mel enhancer sequence that is BLAST-alignable (x-axis) or BLAT-alignable (y-axis) to the pse ortholog.
(D–E) The D. melanogaster (mel) and D. pseudoobscura (pse) orthologs of spa. Left, enhancer diagrams showing known or predicted Su(H), Ets, and Lz/Runx binding sites (red, yellow, and blue, respectively). Right, eye imaginal discs of transgenic mel larvae carrying mel spa-GFP (D) or pse spa-GFP (E). Insets show 24-hour transgenic pupal eyes stained with antibodies against GFP (green) and the cone cell nuclear marker Cut (magenta).