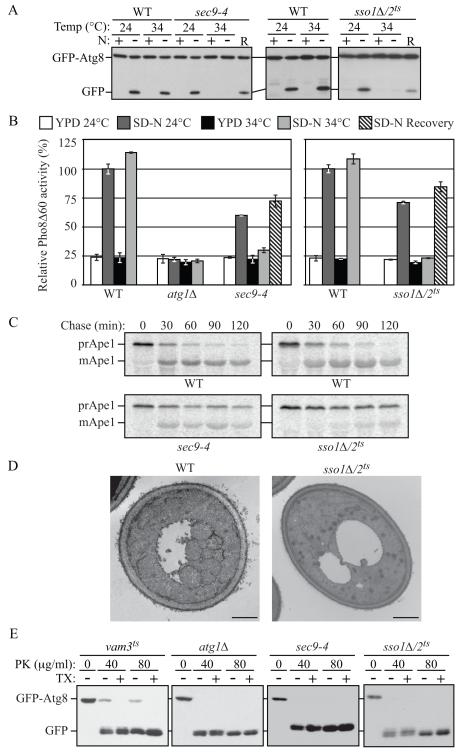
Fig. 2. Both Autophagy and the Cvt Pathway Are Defective in sec9-4 and sso1Δ/2ts Mutants.
(A) The sec9-4 and sso1Δ/2ts (H603) strains and the corresponding wild type (BY4742 and W303) were analyzed with the GFP-Atg8 processing assay at the indicated temperature by nitrogen starvation for 2 h. R, recovery; cells were returned to 24°C after the 34°C incubation for an additional 2 h.
(B) The sec9-4 (JGY236), and sso1Δ/2ts (JGY247) strains and the corresponding wild-type (YTS158 and ZFY202) cells were analyzed for Pho8Δ60 activity before and after starvation for 2 h. The atg1Δ (TYY127) strain was used as a negative control. Error bar, SD from three independent experiments.
(C) The strains from (A) were examined by pulse-chase at NPT and immunoprecipitated with anti-Ape1 antiserum.
(D) Wild-type (pep4Δ vps4Δ; UNY148) or sso1Δ/2ts pep4Δ vps4Δ (UNY142) cells were analyzed by electron microscopy after 1.5 h starvation. Scale bar, 500 nm.
(E) Cultures of vam3ts (UNY162), atg1Δ (TYY164), sec9-4 and sso1Δ/2ts (H603) cells were pre-incubated at 34°C for 30 min, shifted to starvation conditions for 1 h at the NPT and analyzed for sensitivity to proteinase K (PK) with or without 0.2% Triton X-100 (TX) as described in the Supplement. See also Figure S2.