FIGURE 2.
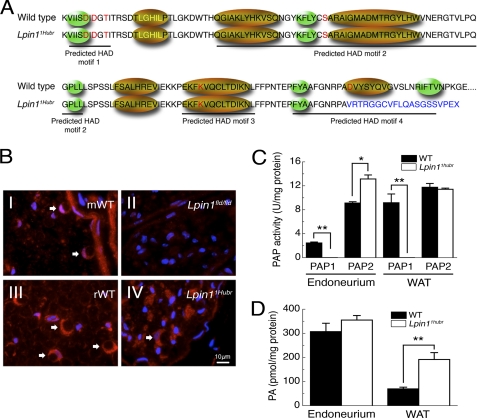
The Lpin1 PAP1 activity is disrupted in Lpin11Hubr rats. A, amino acid sequence showing the four predicted HAD motifs, the conserved amino acids from the HAD family of proteins are indicated in red letters, the transcription coactivator motif LXXIL in yellow letters, green ellipses indicate predicted β-strands, and gold ellipses indicate predicted α-helices (11). The partial intron retention disrupts the conserved amino acid in the predicted HAD motif IV (WT, GNRPAD; Lpin11Hubr, GNRPAV). Moreover, the out-of-frame translation (indicated in blue) disrupts the predicted α-helix and the second predicted β-strand of the predicted HAD motif IV. B, immunolabeling of Lipin 1 (red) and 4′,6-diamidino-2-phenylindole (DAPI; blue) in cross-sections of sciatic nerve derived from wild-type (mWT; B, panel I) or Lpin1fld/fld mice at PND 56 (B, panel II), and derived from wild-type (rWT; B, panel III), or Lpin11Hubr rats (B, panel IV) at PND 90. The endoneurial part of the control nerves shows the expression of Lipin 1 in myelinating Schwann cells (red-stained croissant-shaped cells indicated by white arrows). This labeling is completely absent in Lpin1fld/fld mice but partially preserved in Lpin11Hubr rats. C, PAP1 activity is substantially decreased in sciatic nerve endoneurium and WAT of Lpin11Hubr rats as compared with wild-type rats at PND 21. PAP2 activity is increased in sciatic nerve endoneurium of Lpin11Hubr rats as compared with wild-type rats, whereas no significant difference in WAT PAP2 activity was observed between genotypes (*, p < 0.005; **, p < 0.001; n = 5–6 per group). D, at PND 21, PA levels were unchanged in sciatic nerve and increased in WAT tissue of Lpin11Hubr rats as compared with wild-type rats (*, p < 0.001; n = 6 per group). Data are expressed as mean ± S.E.