Abstract
Pluripotent stem cells possess a tremendous potential for the treatment of many diseases because of their capacity to differentiate into a variety of cell lineages. However, they provide little promise for muscle-related diseases, mainly because of the lack of small molecule inducers to efficiently direct myogenic conversion. Retinoic acid, acting through the retinoic acid receptor (RAR) and retinoid X receptor (RXR), affects stem cell fate determination in a concentration-dependent manner, but it only has a modest efficacy on the commitment of ES cells into skeletal muscle lineage. The RXR is very important for embryonic development but is generally considered to act as a silent partner of RAR in a non-permissive mode. In this study, we have examined whether activation of the RXR by rexinoid or RXR-specific signaling play a role in the specification of stem cells into muscle lineage. Our findings demonstrate that mouse ES cells generate skeletal myocytes effectively upon treatment with rexinoid at the early stage of differentiation and that on a molecular level, rexinoid-enhanced myogenesis simulates the sequential events observed in vivo. Moreover, RXR-mediated myogenic conversion requires the function of β-catenin but not RAR. Our studies establish the feasibility of applying the RXR agonist in cell-based therapies to treat muscle-related diseases. The aptitude of mouse ES cells to generate skeletal myocytes following rexinoid induction also provides a model system to study the convergence of different signaling pathways in myogenesis.
Keywords: Chromatin, Chromatin Immunoprecipitation (ChIP), Differentiation, Gene Regulation, Nuclear Receptors, Stem Cells
Introduction
Skeletal muscle development is a highly ordered process orchestrated by many myogenic regulatory factors (MRFs)2, including Myf5, MyoD, myogenin, and Mrf4 (1). Although Myf5 and MyoD initiate the transcription of muscle-specific genes and direct the cells into skeletal muscle lineage, myogenin and Mrf4 mainly regulate the later stages of development such as the fusion of myoblasts to myotubes (1–3). Although MRFs are able to regulate each other, they require the signaling of early factors to be temporally activated (4). Meox1 and Pax3 are early factors that specify the progenitor cells, whereas MyoD is positively regulated by Pax3 and Myf5 (5–7). Wnt signaling via β-catenin is important for the induction of MRFs and skeletal myogenesis (6).
The temporal expression pattern of MRFs in ES cells reflects the sequential events observed during skeletal myogenesis in vivo (8). Similar to the ES cell system, pluripotent embryonic carcinoma cells respond well to developmental cues in vitro to differentiate into the cell types of all three germ layers (9). The differentiation of embryonic carcinoma stem cells simulates the molecular and cellular processes that occur during ES cell differentiation and early embryonic development (10). The pluripotent stem cells are promising resources for cell-based therapies but have proved difficult to apply in muscle-related diseases, mainly because of the lack of small molecule inducers to effectively direct skeletal myogenic conversion (11).
Retinoic acid (RA) is essential for a broad array of biological processes, including vertebrate body shaping, tissue homeostasis, apoptosis, and cell differentiation (12, 13). High concentrations of RA (>10−7 m) enhance neuronal differentiation but inhibit myogenesis, whereas low concentrations (<10−7 m) enhance myogenic conversion in ES and embryonic carcinoma cells (14–16). The diverse effects of RA are primarily mediated through retinoic acid receptors (RAR), which act as ligand-inducible transcription factors to regulate RA-responsive genes (17). The function of RAR depends on retinoid X receptors (RXR). RAR bind to specific DNA constitutively with the RXR as a heterodimer within the genes they govern and, upon ligand induction, recruit the p300 coactivator complex to activate gene transcription (18, 19). The RAR-RXR dimer binds to consensus sequences, including a DR5 motif, in which ligand induction is through RAR, whereas RXR is generally considered a silent partner (20). In addition to RAR, RXR can dimerize with one-third of the known nuclear receptors, and RXR is amenable to ligand activation in the permissive heterodimers or homodimers (21, 22).
Although RA is the best characterized inducer for myogenic conversion, it only has a modest efficacy on ES cells. Thus, it is imperative to comprehend on a molecular level how different signaling pathways converge to regulate the specification of muscle lineage to find efficient inducers that can produce large quantities of skeletal myocytes. In this study, we have examined the mechanisms of signaling-dependent events during myogenic conversion. Our studies have determined a role for RXR-specific signaling in this process and identified the RXR agonist as an effective inducer for the differentiation of ES cells into skeletal myocytes.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
P19 cells (ATCC) were grown in minimal essential medium α (Invitrogen) supplemented with 5% of fetal bovine serum, 5% of bovine calf serum (PAA), and 1% penicillin/streptomycin. After 4 days of aggregation in Petri dishes, the cells were transferred to tissue culture dishes, and coverslips were coated with 0.1% gelatin and grown for a further 5 days. D3 ES cells (ATCC) were maintained in DMEM (Invitrogen) supplemented with 15% of fetal bovine serum (PAA), 1% of penicillin/streptomycin, 1% of non-essential amino acids (Invitrogen), and 1.18 mm β-mercaptoethanol. Maintenance cultures were supplemented with 1000 units/ml of leukemia inhibitory factor (Chemicon). For differentiation, cells were grown in hanging drops for 48 h after which they were washed into Petri dishes and maintained for a further 5 days in suspension. Cells were then transferred to tissue culture dishes and coverslips or harvested for real-time RT-PCR and Western blotting analysis. D3 terminal differentiation medium was DMEM F12 supplemented with 1% N2 (Invitrogen) and 1% penicillin/streptomycin. RA was from Sigma-Aldrich, bexarotene from LC Laboratories, and Ro 41-5253 from Biomol International.
Immunofluorescence Microscopy
The cells were fixed on coverslips as described previously (23) and incubated with indicated primary antibodies overnight at 4 °C after which they were incubated with fluorescent secondary antibodies followed by incubation with 25 ng/ml of Hoechst (Molecular Probes) for DNA staining. Microscopy analysis was performed with a Zeiss Axiovert 200 m (24). Image acquisition was carried out with an AxioCam HRm monochrome camera. Images captured through different fluorescence filters (488 and 594) were processed and merged by Zeiss AxioVision Rel 4.6 software. Skeletal myocytes were quantified as the percentage of cells staining positive for skeletal markers of the total population of cells. Primary antibodies used were anti-myosin heavy chain MF20 and anti-MyoD (Santa Cruz Biotechnology). Secondary antibodies used were Alexa Fluor 488 goat anti-mouse, Alexa Fluor 488 goat anti-rabbit, and Alexa Fluor 594 donkey anti-mouse (Invitrogen).
ChIP
The cell aggregates were fixed on day 4, sonicated, and immunoprecipitated as described previously (25). A sample of 5% of total chromatin was set aside as input control. For immunoprecipitation, antibody against p300, RXR-α, and RAR-α were from Santa Cruz Biotechnology, and β-catenin was from Millipore. IgG antiserum (Zymed Laboratories, Inc., CA) was used for negative ChIP control. DNA was purified using an Omega Bio-tek Cycle Pure kit, and samples were analyzed using SYBR Green real-time PCR. Primer pairs used for amplification were described previously (26).
Western Blot Analysis
The cell pellets were lysed by incubation in whole cell extract buffer (10% glycerol, 50 mm Tris-HCl (pH 7.6), 400 mm NaCl, 5 mm EDTA, 1 mm DTT, 1 mm PMSF, 1% Nonidet P-40) for 30 min (27). Protein concentrations were quantified by the Bradford Method (Bio-Rad) using a Multiscan spectrum photospectrometer and the Multiscan software (Thermo Scientific). Protein was separated on sodium dodecyl sulfate polyacrylamide gel and transferred to an immunoblot PVDF membrane (Bio-Rad). Membranes were incubated overnight in primary antibody followed by incubation with secondary antibody and visualized using Western Lightning chemiluminescence reagents (PerkinElmer Life Sciences). Quantification was performed using Scion Image (Scion Corp.). Primary antibodies used were anti-p300, RXR-α, RAR-α (Santa Cruz Biotechnology), anti-myogenin (F5D hybridoma), and anti-β-tubulin (E7 hybridoma).
Real-time RT-PCR Analysis
Total RNA was isolated using a Total RNA Kit I according to the manufacture's recommendations (Omega), and reverse-transcribed using a high-capacity cDNA reverse transcription kit (Applied Biosystems) as described previously (28). Real-time PCR was performed using a SYBR Green and ROX PCR Master Mix HotStarTaq DNA polymerase kit (Qiagen) and conducted on the Applied Biosystems 7500 Fast real-time PCR system. The amount of targets, normalized to the GAPDH endogenous reference and relative to calibrator control, was calculated using the arithmetic formula 2--ΔΔCT.
Transfection and Luciferase Assay
Transient transfection was performed with a reporter plasmid by using ExGen 500 as described previously (29). Briefly, P19 aggregates were transfected with a RAR luciferase and a RSV-β-gal reporter and then induced with bexarotene or RA. A luciferase assay was performed according to the manufacturer's recommendations (Promega). Luciferase activities are expressed as fold induction relative to the untreated controls after being normalized to the β-galactosidase activity.
RESULTS
Effects of Rexinoid on Myogenic Conversion of Pluripotent P19 Cells
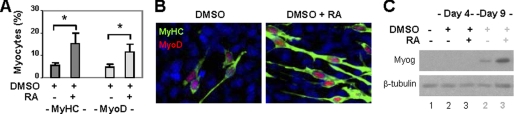
Pluripotent P19 cells are an excellent stem cell system to study cellular differentiation and to identify small molecule inducers for the specification of muscle lineage (9). They can be induced into differentiation with aggregated cultures. However, in the absence of exogenous stimuli or small molecule inducers, aggregation leads only to the expression of markers of the mesoderm but not myoblasts (30). Treatment of aggregates with DMSO induces small percentages of skeletal myocytes, whereas addition of all-trans RA enhances the commitment of muscle lineage (26). As shown in Fig. 1A, DMSO treatment induced about 5% of skeletal myocytes by day 9 of differentiation. In contrast, cotreatment of cells with RA enhanced myogenic conversion to about 15%, as determined by quantitative microscopy of myosin heavy chain and MyoD staining (Fig. 1A). Moreover, MyoD and myosin heavy chain costained to the elongated bipolar myocytes, whereas myogenin protein was also detected by day 9, which is indicative of skeletal myocyte identity (Fig. 1, B and C). Thus, RA signaling is important for the specification and development of skeletal muscle lineage.
FIGURE 1.
Effects of RA on skeletal myogenesis. A, P19 aggregates were treated with DMSO (1%) and RA (10 nm) and costained for myosin heavy chain (MyHC) and MyoD on day 9. Quantification is plotted as fractions of cells differentiated into skeletal myocytes. *, p < 0.05. Error bars are the S.D. of four independent experiments. B, the representative images of myosin heavy chain and MyoD costaining. C, Western analysis of myogenin expression with undifferentiated cells as control. β-tubulin was used as a loading control.
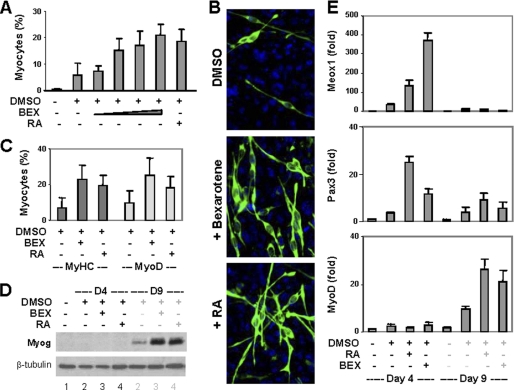
Because RXR is very important for the early stages of embryonic development (31–33), we next used bexarotene, an RXR-selective agonist, to determine the impact of RXR signaling on the specification of muscle lineage. In the presence of DMSO, bexarotene enhanced myogenic conversion in a concentration-dependent manner and achieved an efficacy similar to RA (Fig. 2, A and B). MyoD and myogenin protein were also detected by day 9 of differentiation and expressed at comparable levels to RA treatment (Fig. 2, C and D). In consistence with literature (26), the transcripts of Meox1 and Pax3 were increased by RA on day 4, whereas MyoD increased on day 9 of differentiation (Fig. 2E). Intriguingly, bexarotene caused a greater increase in the Meox1 transcript level than RA, whereas RA caused a larger increase in the Pax3 transcript level than bexarotene (Fig. 2E). The levels of MyoD transcript appeared to reflect the efficacies of myogenic differentiation (Fig. 2E). Thus, the temporal expression pattern of myogenic-specific regulators induced by bexarotene in the P19 model is similar to that seen in vivo, and the RXR agonist is an effective inducer for the commitment of skeletal muscle lineage.
FIGURE 2.
Effects of rexinoid on P19 cell differentiation. A, P19 aggregates were treated with bexarotene (BEX, 1 nm, 10 nm, 100 nm, or 1 μm) or RA (10 nm) in the presence of DMSO and stained for myosin heavy chain on day 9. Quantification is presented as fractions of cells differentiated into skeletal myocytes. Error bars are the S.D. of five independent experiments. B, the representative microscopic images. C, the cells were also costained for MyoD and quantified in comparison with myosin heavy chain (MyHC)-positive cells. D, Western blot analysis of myogenin expression with undifferentiated cells as control. β-tubulin was used as a loading control. E, the relative mRNA levels of Meox1, Pax3, and MyoD were determined by quantitative real-time RT-PCR and plotted as fold difference in relation to untreated day 4 controls after being normalized to GAPDH.
Effects of Rexinoid on the Differentiation of ES Cells into Skeletal Myocytes
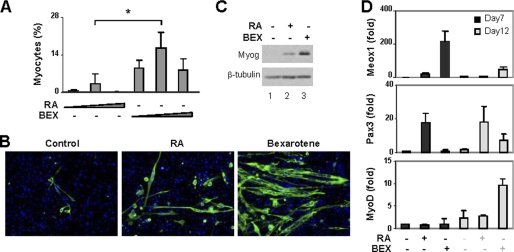
Because ES cells have proved largely unresponsive to RA in the context of myogenic conversion, we next tested rexinoid in this system. A hanging-drop method was used to form embryoid bodies (EB) to induce ES cell differentiation, whereas DMSO was omitted from the protocol because of toxicity to the cells. Different concentrations of bexarotene were used to treat the embryoid bodies, and the development of myoblasts was examined by immunofluorescence microscopy. Consistent with previous reports, RA had a modest efficacy, about 3%, at converting the ES cells into skeletal myocytes (Fig. 3, A and B). In contrast, bexarotene was 5-fold more potent than RA and significantly increased the conversion of myogenic lineage to about 16% (Fig. 3, A and B). Moreover, bexarotene was also more efficient at inducing the expression of myogenin protein (Fig. 3C). As with the P19 cell system, bexarotene increased Meox1 transcripts about 11-fold more than RA, whereas RA was significantly more effective at augmenting Pax3 transcripts (Fig. 3D). Again the levels of MyoD transcripts appeared to reflect the efficacies of myogenic conversion (Fig. 3D). Together these findings demonstrate that the RXR agonist is a more effective inducer than RA to specify ES cells into skeletal muscle lineage.
FIGURE 3.
Effects of rexinoid on ES cell differentiation. A, RA (5, 10, or 20 nm) or bexarotene (BEX, 20, 50, or 100 nm) were used during embryoid body formation. Cells were then plated on coverslips on day 7 and stained for myosin heavy chain and MyoD on day 20. Microscopic analyses were performed and plotted as fractions of cells differentiated into skeletal myocytes. *, p < 0.05. Error bars are the S.D. of four independent experiments. B, representative images of myosin heavy chain staining. C, Western analysis of myogenin with undifferentiated ES cells as control. β-tubulin was used as a loading control. D, the relative transcript levels of Meox1, Pax3, and MyoD were determined by real-time RT-PCR analysis and plotted as fold variance of untreated day 7 controls after being normalized to GAPDH.
Roles of β-Catenin, Meox1, and Pax3 in Rexinoid-enhanced Myogenic Conversion
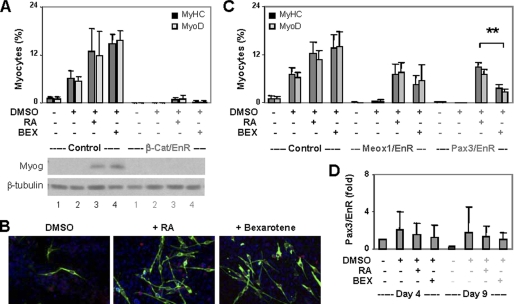
To delineate the molecular pathway of rexinoid action in myogenic differentiation, we next used the P19 cell system to take advantage of several established stable cell lines. RA-enhanced skeletal myogenesis depends on the function of β-catenin (26). To determine the requirement of β-catenin for rexinoid-enhanced myogenesis, we employed a clone of cells stably expressing a dominant negative β-catenin in which the transcriptional activation domain is replaced by an engrailed repressor domain (26). The engrailed repression domain (EN-2) silences gene transcription by interacting with the members of the Groucho/transducin-like enhancer family of transcriptional repressors (4). Cells harboring the empty vector were used as a control. As shown in Fig. 4, A and B, the control cells were differentiated into skeletal myocytes by bexarotene and RA with similar efficacies as the parental cells (compare with Fig. 2A). However, the dominant negative β-catenin cells failed to differentiate into skeletal myocytes regardless of treatments, and myogenin was not detected (Fig. 4A). Therefore, bexarotene, just as RA, cannot bypass the β-catenin pathway to direct skeletal myogenesis, and the function of β-catenin is very important for RXR-mediated myogenic conversion.
FIGURE 4.
Roles of β-catenin, Meox1, and Pax3 in rexinoid-enhanced skeletal myogenesis. A, a clone of P19 cells expressing a dominant negative β-catenin (β-cat/EnR) was differentiated with bexarotene (BEX, 100 nm) or RA (10 nm) and stained for myosin heavy chain (MyHC) and MyoD. Cells harboring the empty vector were used as control. Quantification is plotted as fractions of cells differentiated into skeletal myocytes. Error bars are the S.D. of three independent experiments. Western blot analysis was used to determine myogenin expression with undifferentiated ES cells as control. B, the representative images of myosin heavy chain and MyoD costaining. C, cells expressing the dominant negative Meox1 (Meox1/EnR) or Pax3 (Pax3/EnR) were differentiated as in A. **, p < 0.01. Cells harboring the empty vector were used as control. Error bars are the S.D. of four independent experiments. D, the relative mRNA levels of EnR were determined by quantitative real-time RT-PCR and plotted as fold difference in relation to untreated day 4 controls after being normalized to GAPDH.
Next, we employed Meox1 and Pax3 dominant negative cells (34, 35) to study the roles of Meox1 and Pax3 in rexinoid-enhanced myogenesis. As shown in figure 4C, DMSO did not convert these cells into myoblasts, in consistence with previous reports (34, 35). However, both bexarotene and RA induced the differentiation of the Meox1 and Pax3 dominant negative cells into skeletal myocytes, albeit with lower efficacies (Fig. 4C). Intriguingly, although the levels of engrailed Pax3 transcripts were comparable in the bexarotene- and RA-treated cells, the dominant negative Pax3 appeared to be less detrimental to the RA-induced myogenic conversion (Fig. 4, C and D). This may be due to RA augmenting the endogenous Pax3 expression more effectively than bexarotene (Fig. 2E) to titrate out the interference of dominant negative Pax3. Taken together, these data suggest that rexinoid directs skeletal myogenesis through molecular pathways distinct from RA.
RAR-independent RXR Signaling in Myogenic Conversion
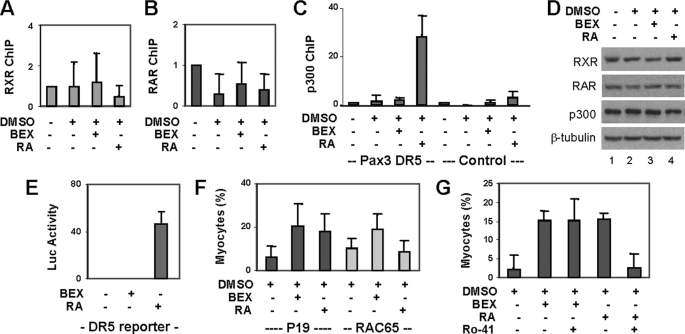
A RAR binding region has been identified previously at the Pax3 locus (26). This region contains a DR5 motif, two direct repeats of the consensus sequence (5′-PuGGTCA) separated by five nucleotides, for occupancy by a RAR-RXR heterodimer (36, 37). We therefore examined the binding of RAR and RXR to this Pax3 locus. ChIP analysis showed that both RAR and RXR occupy this region constitutively (Fig. 5, A and B), consistent with the binding model of the DR5 motif (25). The recruitment of transcriptional coactivator p300 to this region, however, was only augmented by the addition of RA but not bexarotene, although the levels of p300 protein in these cells were constant (Fig. 5, C and D). Thus, RXR possibly acts as a silent partner of RAR at this locus to augment Pax3 gene expression. We also performed a ChIP analysis with a β-catenin antibody to examine the specificity of the ChIP protocol and did not detect apparent β-catenin binding to this locus, regardless of treatment in comparison to the IgG-negative ChIP control (n = 4).
FIGURE 5.
Role of RAR in RXR-specific signaling for myogenic conversion. A, P19 aggregates were treated with bexarotene (BEX, 100 nm) or RA (10 nm). RXR binding to the Pax3 locus was examined by ChIP analysis on day 4 of differentiation. Input DNA was used as internal control. Error bars are the S.D. of four independent experiments. B, RAR binding was analyzed as in A. C, occupancy by p300 at the Pax3 locus was also analyzed. A set of primer that does not cover the DR5 motif was used in parallel as control. D, Western blot analysis of RAR, RXR, and p300 protein. E, P19 aggregates were transfected with a DR5 luciferase reporter (0.1 μg) and a RSV-β-Gal (0.2 μg), induced with bexarotene or RA, and assayed for luciferase activities. Values are the fold induction compared with the untreated control with β-gal as internal controls. F, RAC65 cells were differentiated and analyzed by immunofluorescence microscopy. Quantification is plotted as fraction of cells differentiated into skeletal myocytes. Error bars are the S.D. of four independent experiments. G, P19 aggregates treated with bexarotene or RA were also cotreated with a RAR-selective antagonist Ro 41-5253 (10 nm). Quantification is plotted as fractions of cells differentiated into skeletal myocytes. Error bars represent the S.D. of three independent experiments.
We next used a reporter approach to examine the effects of rexinoid on transactivation requiring RXR acting as a silent partner of RAR. The cells were transfected with a well characterized DR5 RA-responsive reporter (38) during aggregation and treated with bexarotene or RA. As shown in Fig. 5E, RA, but not bexarotene, was able to transactivate the reporter. These data demonstrate that rexinoid does not affect the binding motif for a RAR-RXR heterodimer in the P19 differentiation system, suggesting an additional role for RXR in the specification of skeletal muscle lineage, acting through an activated or permissive mode.
To delineate whether RAR is required for rexinoid-enhanced skeletal myogenesis, we used RAC65 cells, which contain a dominant negative RAR-α that effectively blocks the DNA binding of wild-type receptors (39, 40). These cells are non-responsive to the RAR agonist but respond to the RXR agonist for neuronal differentiation (41, 42). As shown in figure 5F, bexarotene, but not RA, enhanced the specification of skeletal muscle lineage in the RAC65 cells. To further examine the role of RAR in rexinoid-enhanced myogenic conversion, we also used Ro 41-5253, a RAR-specific antagonist (43). As shown in Fig. 5G, Ro 41-5253 at 10 nm concentration selectively inhibited RA-enhanced skeletal muscle development, whereas bexarotene-enhanced myogenesis was not compromised. Thus, RXR directs the specification of skeletal muscle lineage through mechanisms independent of RAR.
DISCUSSION
We have examined whether the activation of RXR by rexinoid or RXR-specific signaling plays a role in myogenic differentiation. Our findings show that mouse ES cells can effectively generate skeletal myocytes following induction with the RXR agonist at the early stage of differentiation and that the molecular pathways involved in rexinoid-directed skeletal myogenesis recapitulate the sequential events observed in vivo. Furthermore, rexinoid-enhanced myogenic conversion is mediated in a RAR-independent manner. Our studies establish the feasibility of applying rexinoid in stem cell therapies, particularly exploring RXR-specific signaling to convert ES cells into skeletal muscle lineage. The aptitude of ES cells to generate myocytes in response to rexinoid also offers a model system to decipher the complex signaling cascade implicated in skeletal muscle development and to develop non-toxic protocols for generating large quantities of skeletal myocytes.
Pluripotent stem cells, regardless of their origin, possess the potential of developing into skeletal myocytes, among many other cell lineages (44). The central issue is how to control a specific signaling pathway to preferentially enhance myogenic conversion in an efficacy suitable for clinical therapies. RA is able to enhance skeletal myogenesis in pluripotent EC stem cells if used in combination with other small molecular inducers such as DMSO (Fig. 1). However, DMSO is toxic to ES cells, and RA alone has only a modest effect on the differentiation of ES cells into myogenic lineage (Fig. 3). We found that rexinoid is a more effective inducer than RA for skeletal myogenesis in the ES cell system (Fig. 3). This is significant because to date there has been very little success at directing ES cells into skeletal muscle lineage, and thus, no methods are currently available to generate a sufficient population of skeletal myocytes for potential cell-based therapies (45).
In addition, our studies also shed new light on the role of RXR in skeletal myogenesis. It is well known that RXR is important for development, and RXR is generally considered to act as the silent partner of RAR in a non-permissive mode (13, 31–33). Our studies establish a role of RXR-specific signaling in the differentiation of stem cells into skeletal muscle lineage, which is independent of RAR and distinct from the action mode of the RAR-RXR heterodimer (Fig. 5). Although the early specification events differ between rexinoid- and RA-enhanced myogenesis, both inducers critically depend on the function of β-catenin, which is required for MyoD expression (Fig. 4).
It appears that rexinoid has a significant impact on an early differentiation marker, Meox1, whereas RA may require DMSO cotreatment to activate Meox1 (Figs. 2 and 3). Our studies have not investigated other genomic targets activated by RXR-specific signaling besides the known MRFs. Additional systems studies are needed to uncover novel signaling pathways involved in rexinoid-enhanced skeletal myogenesis and to determine why the RXR agonist is a more suitable inducer for the ES cell system. Understanding the molecular mechanisms of myogenic specification is very important for manipulating stem cell fate determinations in cell-based therapies. We have identified a potent inducer for a non-toxic protocol to direct the specification of muscle lineage. The efficacy with which rexinoid is able to convert ES cells into skeletal myocytes suggests a potential to extend this strategy to human ES cells and to other types of pluripotent stem cells in view of generating functional skeletal myocytes.
Acknowledgements
We thank Dr. L. Wang for the support on ES cultures; Dr. I. S. Skerjanc for the dominant negative β-catenin, Meox1, and Pax3 stable cell lines; Dr. A. Blais for the MF20 and E7 hybridoma lines; and Dr. B. Jasmin for the F5D line.
This work was supported by an operating grant from the Natural Sciences and Engineering Research Council of Canada and by a graduate scholarship award from the Canadian Institutes of Health Research (to M. L.).
- MRF
- myogenic regulatory factor
- RA
- retinoic acid
- RAR
- retinoic acid receptor(s)
- RXR
- retinoid X receptor(s)
- DMSO
- dimethyl sulfoxide.
REFERENCES
- 1. Tapscott S. J. (2005) Development 132, 2685–2695 [DOI] [PubMed] [Google Scholar]
- 2. Weintraub H., Tapscott S. J., Davis R. L., Thayer M. J., Adam M. A., Lassar A. B., Miller A. D. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 5434–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Francetic T., Li Q. (2011) Transcription 2, 109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen G., Courey A. J. (2000) Gene 249, 1–16 [DOI] [PubMed] [Google Scholar]
- 5. Williams B. A., Ordahl C. P. (1994) Development 120, 785–796 [DOI] [PubMed] [Google Scholar]
- 6. Petropoulos H., Skerjanc I. S. (2002) J. Biol. Chem. 277, 15393–15399 [DOI] [PubMed] [Google Scholar]
- 7. Tajbakhsh S., Rocancourt D., Cossu G., Buckingham M. (1997) Cell 89, 127–138 [DOI] [PubMed] [Google Scholar]
- 8. Rohwedel J., Maltsev V., Bober E., Arnold H. H., Hescheler J., Wobus A. M. (1994) Dev. Biol. 164, 87–101 [DOI] [PubMed] [Google Scholar]
- 9. Yu J., Thomson J. A. (2008) Genes Dev. 22, 1987–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rudnicki M. A., Reuhl K. R., McBurney M. W. (1989) Development 107, 361–372 [DOI] [PubMed] [Google Scholar]
- 11. Allsopp T. E., Bunnage M. E., Fish P. V. (2010) Med. Chem. Commun. 1, 16–29 [Google Scholar]
- 12. Berbis P. (2010) Ann. Dermatol. Venereol. 137, S97-S103 [DOI] [PubMed] [Google Scholar]
- 13. Mark M., Ghyselinck N. B., Chambon P. (2009) Nucl. Recept. Signal 7, e002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jones-Villeneuve E. M., McBurney M. W., Rogers K. A., Kalnins V. I. (1982) J. Cell Biol. 94, 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sato Y., Heuckeroth R. O. (2008) Dev. Biol. 320, 185–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hescheler J., Fleischmann B. K., Lentini S., Maltsev V. A., Rohwedel J., Wobus A. M., Addicks K. (1997) Cardiovasc. Res. 36, 149–162 [DOI] [PubMed] [Google Scholar]
- 17. Niederreither K., Dollé P. (2008) Nat. Rev. Genet. 9, 541–553 [DOI] [PubMed] [Google Scholar]
- 18. Lonard D. M., O'Malley B. W. (2007) Mol. Cell 27, 691–700 [DOI] [PubMed] [Google Scholar]
- 19. Hermanson O., Glass C. K., Rosenfeld M. G. (2002) Trends Endocrinol. Metab. 13, 55–60 [DOI] [PubMed] [Google Scholar]
- 20. Kurokawa R., DiRenzo J., Boehm M., Sugarman J., Gloss B., Rosenfeld M. G., Heyman R. A., Glass C. K. (1994) Nature 371, 528–531 [DOI] [PubMed] [Google Scholar]
- 21. Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schütz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. (1995) Cell 83, 835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gampe R. T., Jr., Montana V. G., Lambert M. H., Wisely G. B., Milburn M. V., Xu H. E. (2000) Genes Dev. 14, 2229–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen J., Halappanavar S., Th' ng J. P., Li Q. (2007) Epigenetics 2, 92–99 [DOI] [PubMed] [Google Scholar]
- 24. St-Germain J. R., Chen J., Li Q. (2008) Epigenetics 3, 342–349 [DOI] [PubMed] [Google Scholar]
- 25. Higazi A., Abed M., Chen J., Li Q. (2011) Epigenetics 6, 202–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kennedy K. A., Porter T., Mehta V., Ryan S. D., Price F., Peshdary V., Karamboulas C., Savage J., Drysdale T. A., Li S. C., Bennett S. A., Skerjanc I. S. (2009) BMC Biol. 7, 67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen J., Ghazawi F. M., Bakkar W., Li Q. (2006) Mol. Cancer 5, 71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen J., Ghazawi F. M., Li Q. (2010) Epigenetics 5, 509–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen J., Halappanavar S. S., St-Germain J. R., Tsang B. K., Li Q. (2004) Cell. Mol. Life Sci. 61, 1675–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McBurney M. W. (1993) Int. J. Dev. Biol. 37, 135–140 [PubMed] [Google Scholar]
- 31. Sapin V., Dollé P., Hindelang C., Kastner P., Chambon P. (1997) Dev. Biol. 191, 29–41 [DOI] [PubMed] [Google Scholar]
- 32. Sucov H. M., Dyson E., Gumeringer C. L., Price J., Chien K. R., Evans R. M. (1994) Genes Dev. 8, 1007–1018 [DOI] [PubMed] [Google Scholar]
- 33. Kastner P., Grondona J. M., Mark M., Gansmuller A., LeMeur M., Decimo D., Vonesch J. L., Dollé P., Chambon P. (1994) Cell 78, 987–1003 [DOI] [PubMed] [Google Scholar]
- 34. Petropoulos H., Gianakopoulos P. J., Ridgeway A. G., Skerjanc I. S. (2004) J. Biol. Chem. 279, 23874–23881 [DOI] [PubMed] [Google Scholar]
- 35. Ridgeway A. G., Skerjanc I. S. (2001) J. Biol. Chem. 276, 19033–19039 [DOI] [PubMed] [Google Scholar]
- 36. Tanaka T., De Luca L. M. (2009) Cancer Res. 69, 4945–4947 [DOI] [PubMed] [Google Scholar]
- 37. Umesono K., Evans R. M. (1989) Cell 57, 1139–1146 [DOI] [PubMed] [Google Scholar]
- 38. Chen J., St-Germain J. R., Li Q. (2005) Mol. Cell. Biol. 25, 525–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pratt M. A., Kralova J., McBurney M. W. (1990) Mol. Cell. Biol. 10, 6445–6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Costa S. L., McBurney M. W. (1996) Exp. Cell Res. 225, 35–43 [DOI] [PubMed] [Google Scholar]
- 41. Jones-Villeneuve E. M., Rudnicki M. A., Harris J. F., McBurney M. W. (1983) Mol. Cell. Biol. 3, 2271–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yokota Y., Ohkubo H. (1996) Exp. Cell Res. 228, 1–7 [DOI] [PubMed] [Google Scholar]
- 43. Wang X. J., Hayes J. D., Henderson C. J., Wolf C. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19589–19594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hirst C. E., Ng E. S., Azzola L., Voss A. K., Thomas T., Stanley E. G., Elefanty A. G. (2006) Dev. Biol. 293, 90–103 [DOI] [PubMed] [Google Scholar]
- 45. Darabi R., Gehlbach K., Bachoo R. M., Kamath S., Osawa M., Kamm K. E., Kyba M., Perlingeiro R. C. (2008) Nat. Med. 14, 134–143 [DOI] [PubMed] [Google Scholar]