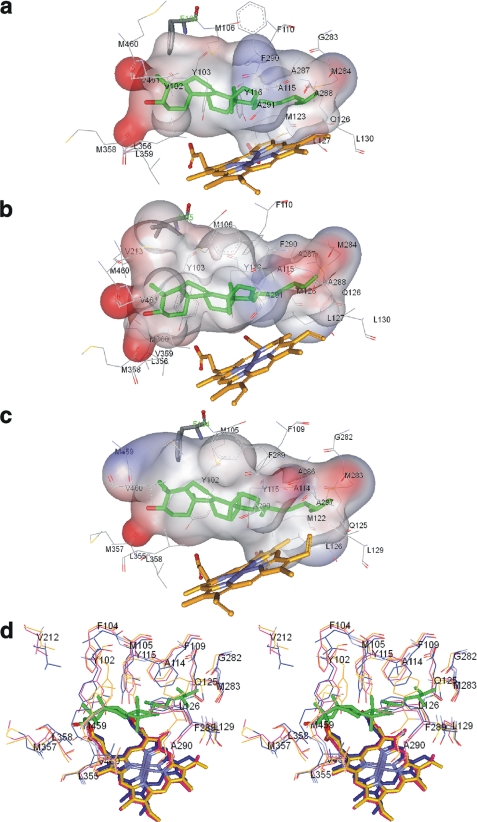
FIGURE 7.
CYP51 substrate binding cavities. The ligand binding surface includes protein atoms located within 4 Å from Ebr modeled into the CYP51 active site. a, T. brucei. Surface volume is 1,433 Å3, and the surface area is 866 Å2. b, T. cruzi. The surface volume is 1,560 Å3, and the surface area is 1016 Å2. c, L. infantum. The surface volume is 1,524 Å3, and the surface area is 934 Å2. Ebr (green), heme, and the substrate preference-defining residues (Phe/Ile) are presented as sticks, and the other residues located within van der Waals contacts are shown as lines. The surface is colored by electrostatic potential. d, stereo view of the active sites residues in the superimposed protozoan CYP51 structures. Magenta, L. infantum; blue, T. cruzi; orange, T. brucei. L. infantum CYP51 numbering is used. In T. cruzi CYP51, in addition to Ile-105 (Phe-104 in L. infantum), there is also a Val instead of Leu in the β1–4 strand (Leu-358 in L. infantum). All other residues in the three CYP51 orthologs are identical.