Abstract
The oxidation and toxicity of dopamine is believed to contribute to the selective neurodegeneration associated with Parkinson disease. The formation of reactive radicals and quinones greatly contributes to dopaminergic toxicity through a variety of mechanisms. The physiological metabolism of dopamine to 3,4-dihydroxyphenylacetaldehyde (DOPAL) via monoamine oxidase significantly increases its toxicity. To more adequately explain this enhanced toxicity, we hypothesized that DOPAL is capable of forming radical and quinone species upon oxidation. Here, two unique oxidation products of DOPAL are identified. Several different oxidation methods gave rise to a transient DOPAL semiquinone radical, which was characterized by electron paramagnetic resonance spectroscopy. NMR identified the second oxidation product of DOPAL as the ortho-quinone. Also, carbonyl hydration of DOPAL in aqueous media was evident via NMR. Interestingly, the DOPAL quinone exists exclusively in the hydrated form. Furthermore, the enzymatic and chemical oxidation of DOPAL greatly enhance protein cross-linking, whereas auto-oxidation results in the production of superoxide. Also, DOPAL was shown to be susceptible to oxidation by cyclooxygenase-2 (COX-2). The involvement of this physiologically relevant enzyme in both oxidative stress and Parkinson disease underscores the potential importance of DOPAL in the pathogenesis of this condition.
Keywords: Electron Paramagnetic Resonance (EPR); Metabolism; Neurological Diseases; Neurotransmitters; Quinones; 3,4-Dihydroxyphenylacetaldehyde; Dopamine; Free Radical
Introduction
Parkinson disease (PD)2 involves specific loss of dopaminergic nuclei in the substantia nigra of the brain (1). Although the exact causes of this selective degeneration are unknown (2), the role of affected cells as centers of dopamine (DA) synthesis, storage, and metabolism suggests that DA may be an endogenous neurotoxin (3, 4) that contributes to the pathogenesis of PD. DA may act as a source of cellular oxidative stress and is known to undergo oxidation (Scheme 1) to cytotoxic radicals and quinones (5–8). Such oxidations can occur spontaneously or via metal- or enzyme-catalyzed mechanisms (9). One-electron oxidation of DA produces a radical capable of interfering with DA storage and causing oxidative protein and DNA modifications (5, 6, 10). Similarly, two-electron oxidation of DA to an ortho-quinone results in reactivity with cellular nucleophiles such as thiols and proteins (8, 11). Both species are capable of redox cycling, which could deplete cellular oxidative defenses. Also, in the presence of transition metals and/or O2, such oxidations could result in the production of ROS capable of inducing lipid peroxidation and damage to other cellular macromolecules (12). Another potential mechanism of toxicity for DA is its physiological metabolism to 3,4-dihydroxyphenylacetaldehyde (DOPAL) (13).
SCHEME 1.
Oxidation of dopamine.
DOPAL is a very reactive aldehyde that is 100–1000-fold more toxic than DA both in vivo and in vitro (14, 15). Physiological levels of DOPAL in the substantia nigra are ∼2 μm; levels as low as 6 μm can exert significant toxicity (16). Reactivity with proteins, presumably via Schiff base formation, is an important mechanism of toxicity for DOPAL (17, 18). Protein modification can result in inhibition of enzyme activity and loss of function for cellular proteins. Other toxicity mechanisms such as mitochondrial dysfunction (19), hydroxyl radical production (20), and interference with DA storage (21) have also been implicated in vitro. However, these mechanisms do not fully account for the high levels of toxicity associated with DOPAL. Therefore, we and others have hypothesized that DOPAL may be capable of undergoing oxidation to a quinone (16, 17, 20, 22).
Oxidation of DOPAL to an ortho-quinone is predicted to have deleterious effects on cells and could result in additional mechanisms of toxicity (16, 17, 20, 22). Work from our laboratory suggests that protein reactivity associated with DOPAL is partially mediated via catechol oxidation (18). Similarly, oxidation of DOPAL to a radical could help explain its high toxicity. However, direct investigations into the oxidation of DOPAL to either a radical or a quinone have not been reported.
In this study, we are the first to report the novel and systematic oxidation of DOPAL to a semiquinone radical and an ortho-quinone. Following UV-visible spectrophotometric identification of two unique oxidation products of DOPAL, spin stabilization electron paramagnetic resonance spectroscopy (EPR) was used to characterize a DOPAL semiquinone radical, whereas two-dimensional NMR experiments were used to identify the structure of the DOPAL quinone. DOPAL was also shown to undergo carbonyl hydration in aqueous media; the quinone seems to exist exclusively in this form. Furthermore, oxidation of DOPAL resulted in enhanced reactivity with protein and ROS production. Also, DOPAL is shown here for the first time to be a substrate for cyclooxygenase-2 (COX-2), providing a potential mechanism for in vivo formation of the oxidized products identified here.
EXPERIMENTAL PROCEDURES
Chemicals
DOPAL was synthesized from epinephrine by the method of Fellman (23) with a slight modification in 20% yield. Purity was 97–99% as determined by HPLC analysis as described previously (18, 24). Synthesized DOPAL was used without further purification. The 13C NMR spectrum of DOPAL is reported here for the first time (internally referenced to DMSO-d6 at 39.51 ppm): δ 200.7 (CHO), 145.4 (C-OH, aromatic), 144.4 (C-OH, aromatic), 123.0 (C, aromatic), 120.4 (CH, aromatic), 117.0 (CH, aromatic), 115.8 (CH, aromatic), and 49.0 (CH2). 1H NMR spectra were collected in both CDCl3 and DMSO-d6 for direct comparison with conflicting reports from the literature (25, 26). Our results are in good agreement with Tsukamoto et al. (26).
Water and buffers used in experiments were treated with Chelex 100 to remove opportune transition metals (27). NaH2PO4 and Tricine were from Research Products International (Mount Prospect, IL), and Cambridge Isotope Laboratories (Andover, MA) provided deuterated solvents and trimethylsilylpropanoate-d4. Human recombinant COX-2 (EC 1.14.99.1; 10,636.25 units/mg) and arachidonic acid were from Cayman Chemicals (Ann Arbor, MI), and DMSO, K3Fe(CN)6, and perchloric acid were from Thermo Fisher Scientific. All other chemicals were from Sigma-Aldrich.
UV-visible Characterization of Oxidized DOPAL
A Hewlett-Packard 8453 diode array spectrometer (standard settings) was used to obtain spectra and λmax of DOPAL (0.5 mm) oxidized by tyrosinase (45 units) in 25 mm NaH2PO4, pH 7.4, over 60 min. Samples (1.5 ml) in quartz cuvettes were stirred with a small stir bar to ensure proper mixing for all readings, and temperature (25 °C) was maintained with a water jacket. Buffer with heat-denatured (80 °C, 5 min) tyrosinase served as a blank, and time-dependent absorbance maxima (λmax) were analyzed versus time to monitor product formation and disappearance. Results are representative of at least three trials.
EPR Spectroscopy
A Bruker EMX spectrometer with an Aqua-X sample holder and high sensitivity cavity equipped with a microwave frequency counter was used for EPR. Sample volumes were 1.0 ml, and spectra were recorded at room temperature. Typical EPR parameters were as follows: 3510.75 G center field; 24 G sweep width; 9.854 GHz microwave frequency; 20 milliwatt power; 2.52 × 104 receiver gain; 100 kHz modulation frequency; 0.20 G modulation amplitude; 40.96 ms conversion time; 81.92 ms time constant; 41.93 s sweep time; and up to 10 X-scans for each 1024-point spectrum. The resultant spectra are representative of at least three trials. Relative radical concentrations were determined by comparing double integrations (using WinEPR) with a stable radical (2,2,5,5-tetramethyl-1-pyrrolidinyloxy-3-carboxylic acid) of known concentration (data not shown).
Spin stabilization (28, 29) experiments were carried out with Mg2+ (0.5 m MgCl2 in 25 mm Tricine buffer, pH 7.4) to visualize the semiquinone radical. DOPAL (400 μm) was oxidized enzymatically with either 10 units of tyrosinase or 10 milliunits of HRP/10 μm H2O2 or chemically with either 0.25 eq of NaIO4 or 0.5 eq of K3Fe(CN)6. For comparison, DA and 3,4-dihydroxyphenylacetic acid (DOPAC) were also oxidized by tyrosinase under spin stabilization conditions.
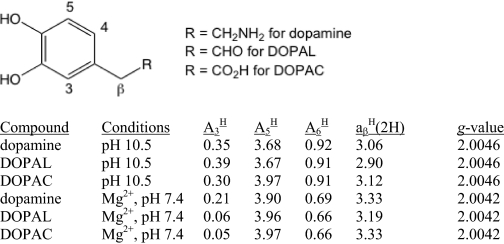
High pH (40 mm NaH2PO4 pH 10.5) was used to induce auto-oxidation (30) of the catechol substrates (DOPAL, DOPAC, and DA, 400 μm each) under non-spin-stabilizing conditions (i.e. without Mg2+). The spectra were simulated with WinSim and WinSim 2002 software (31) to determine the correlation coefficients and splitting constants. DOPAL (400 μm in 0.5 m MgCl2, 25 mm Tricine buffer, pH 7.4) was also incubated with 2.0 eq of H2O2 to test the contribution of H2O2 to DOPAL oxidation. The g values for DA, DOPAL, and DOPAC were determined using 1,4-benzosemiquinone as a standard (g = 2.00469 (in aqueous solution)) (32).
NMR Spectroscopy
All products were characterized using Avance 300, Avance 400, and Avance 600 Bruker NMR spectrometers (Billerica, MA) operating at 300, 400, and 600 MHz with quadranuclei, broadband observe, and broadband inverse probes, respectively. The spectra in D2O were referenced to trimethylsilylpropanoate-d4 (0.0 ppm), whereas samples in 1:4 D2O/acetone-d6 were referenced internally to acetone-d6 (2.05 ppm).
For initial oxidation experiments, DOPAL in D2O (∼20 mm) was carefully oxidized with 1.00 eq of NaIO4 (stock solution dissolved in D2O), and transferred into an NMR tube. 1H spectra were then collected. The reaction was repeated with 10 mm NaH2PO4 (pD 7.5 in D2O), and with a 4-fold excess of acetone-d6. For structure determination experiments, the reaction mixture was diluted 1:4 with acetone-d6 (prechilled on dry ice) 60 s after the addition of NaIO4, and then transferred into an NMR tube and immediately analyzed at −20 °C. The oxidation product was characterized via a variety of one- and two-dimensional homonuclear and heteronuclear experiments (1H, 1H-1H correlated spectroscopy (COSY), 1H-1H nuclear Overhauser effect spectroscopy (NOESY), 1H-13C heteronuclear multiple quantum correlation (HMQC), and 1H-13C heteronuclear multiple bond correlation (HMBC)). All two-dimensional experiments used gradient-assisted pulse sequences, inverse detection, and a relaxation delay of 4.0 s. Typical experimental parameters were as follows: 1H (f1, 12.0 ppm; TD, 32k; NS, 16; DS, 4); COSY (f1 and f2, 10.0 ppm; TD, 4k; TD1, 400; NS, 16; DS, 128); NOESY (f1, 12.2 ppm; TD, 2k; TD1, 200; NS, 16; DS, 32; mixing times, 1.75, 1.15, and 0.55); 13C (f1, 240.0 ppm; TD, 32k; NS, 4000); 13C-1H HMQC (f1[13C], 220.8 ppm; f2[1H], 10.0 ppm; TD, 2k; TD1, 256; NS, 24; DS, 128); and 13C-1H HMBC (f1[13C], 254.8 ppm; f2[1H], 10.0 ppm; TD, 4k; TD1, 400; NS, 40; DS, 128). TD, NS, and DS refer to time domain data points, number of scans, and dummy scans, respectively, and f1 and f2 refer to sweep width. NMR data were processed with TOPSPIN 1.3 software. One-dimensional 1H data were processed with zero-filling to 64,000 data points and 0.2 Hz exponential line broadening, whereas two-dimensional NMR data were processed with zero-filling to 4096 points in acquisition and 1024 points in second dimension.
The formation of a DOPAL hydrate was evident from the spectra collected in D2O prior to oxidation. For determination of the Keq of hydration, stock DOPAL solutions (8, 20, and 24 mm) were prepared from three different synthetic batches in both D2O and 10 mm NaH2PO4 buffer (pD 7.5 in D2O). 1H spectra were collected, and integration ratios (hydrate:aldehyde) for all protons were calculated for each sample on three different days.
SDS-PAGE Protein Cross-linking
Oxidized or non-oxidized DOPAL (50 μm) was reacted with glyceraldehyde 3-phosphate dehydrogenase (GAPDH, 0.3 mg/ml) in 50 mm NaH2PO4, pH 7.4, at 37 °C for 4 h and then analyzed via SDS-PAGE (4% stacking, 10% resolving gels, and 9 μg of protein loaded/well) and densitometry (NIH ImageJ software) as described previously (18). DOPAL oxidized by various methods (1.0 eq of NaIO4, 36 units/ml tyrosinase, and 2.0 eq of K3Fe(CN)6) was added to the protein at specific time points (10 or 60 s following oxidation). The resultant protein reactivity and cross-linking were indicated by a decrease in intensity of the control protein band with corresponding formation of higher molecular weight species. The results are representative of two separate trials.
DOPAL Auto-oxidation and ROS Production
Superoxide formation during DOPAL auto-oxidation was investigated using the method of Eyer (30, 33). UV-visible spectrophotometry (kinetic settings) was used to measure the change in absorbance at 400 nm during auto-oxidation of DOPAL (0.5 mm) in 50 mm NaH2PO4, pH 7.8. The reaction was repeated in the presence of SOD (8 μm, 1200 units/ml; or 40 μm, 6000 units/ml). The samples (1.5 ml) were stirred at 37 °C and blanked against buffer with or without SOD. The readings were taken every 3 min for 12 h, and the reported results are representative of at least three runs.
Oxidation of DOPAL by COX-2
The initial linear slope and kcat of DOPAL (50 μm) oxidized by human recombinant COX-2 (100 units/ml) were determined by measuring the formation of oxidized products and the disappearance of DOPAL over time. The assay conditions of Wangpradit et al. (34) were utilized for all reactions (100 mm KH2PO4, pH 7.4, 100 μm potassium arachidonate, 1 μm hematin, 1 ml volume). 100-μl aliquots were removed at various time points, treated with perchloric acid (6 μl), centrifuged (10,000 × g, 5 min), and analyzed quantitatively with an Agilent 1200 series capillary system HPLC with photodiode array detector. The concentration of DOPAL (non-oxidized) was determined by the calibration curve method described previously (18, 24). To estimate the concentrations of oxidized DOPAL in each aliquot, a calibration curve (not shown) was constructed using 50 μm DOPAL oxidized by various concentrations of NaIO4. This allowed the correlation of the total HPLC peak area with the concentration of oxidized DOPAL.
The amounts of DOPAL oxidized by COX-2 (aliquots at 0-, 1.5-, and 5-min time points) were plotted to determine the initial linear slope; kcat was then calculated by accounting for the amount of enzyme used in each assay and the molecular mass of COX-2 (72 kDa). The assay was repeated three times for DOPAL and once with DA (65 μm) for comparison. Also, a trial, with the enzyme omitted but all other components present, was utilized to determine base-line (non-enzymatic) DOPAL oxidation.
RESULTS
UV-visible Characterization
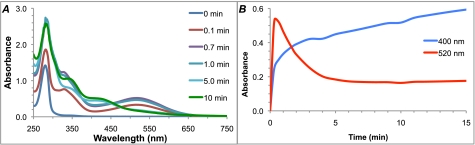
Rearrangement of DOPAL upon oxidation was evident colorimetrically. The treatment of DOPAL with tyrosinase catalyzed the stepwise formation of two unique chromophores, an early and transient pink species followed by a relatively more stable yellow species. UV-visible composite scans (Fig. 1A) allowed the correlation of the early transient species with a λmax of 520 nm and the later species with a λmax of 400 nm. The relative intensity of these species over time is demonstrated graphically in Fig. 1B. Oxidation of DOPAL with NaIO4 followed a similar course, although it occurred too quickly to capture changes spectroscopically (not shown). Because of the ability of tyrosinase and NaIO4 to oxidize DA to a quinone (7), we expected DOPAL to undergo a similar oxidation. However, the λmax of the initial species was higher than expected for a quinone (35). Its transient nature and the susceptibility of quinones to undergo comproportionation (30) in the presence of their reduced counterparts (i.e. hydroquinones and catechols) suggested that the pink species was a radical (Scheme 2).
FIGURE 1.
UV-visible spectrophotometric characterization of DOPAL oxidation products. A, overlaid spectra of DOPAL oxidized by tyrosinase at pH 7.4 show the formation of two unique species. The early oxidation product is transient and centered at 520 nm, whereas the later species is relatively more stable and centered at 400 nm. Only the first several minutes are displayed for clarity. B, relative absorbance of the two oxidation products over time.
SCHEME 2.
Comproportionation of quinone and catechol to radicals.
EPR Spectroscopy
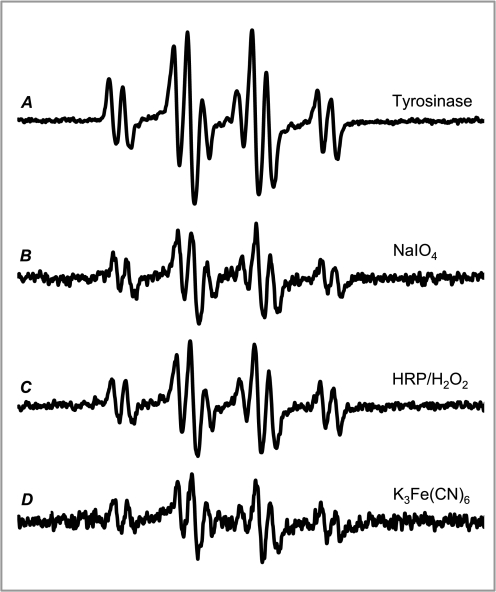
The use of redox-inactive divalent metal cations, known to stabilize semiquinone radicals (28, 29), allowed the spectra of a DOPAL semiquinone radical to be collected for the first time. Both two-electron (tyrosinase and NaIO4) and one-electron oxidizing agents (HRP/H2O2 and K3Fe(CN)6) produced the same DOPAL semiquinone radical as seen by identical EPR spectra (Fig. 2). Regardless of the oxidation method, the radical levels were below the detection limit at pH 7.4 in the absence of Mg2+.
FIGURE 2.
EPR spin stabilization spectra of the DOPAL semiquinone radical formed by a variety of methods. DOPAL oxidized by tyrosinase (A), NaIO4 (B), HRP-H2O2 (C), and K3Fe(CN)6 (D) in the presence of Mg2+ resulted in the formation of a semiquinone radical. The overall efficacy of oxidation methods for radical production under the experimental conditions utilized was tyrosinase > HRP-H2O2 > NaIO4 > K3Fe(CN)6. For both the enzymatic (A and C) and chemical (B and D) methods, two-electron oxidation and comproportionation (A and B) appeared to produce higher levels of the radical than direct one-electron oxidation (C and D).
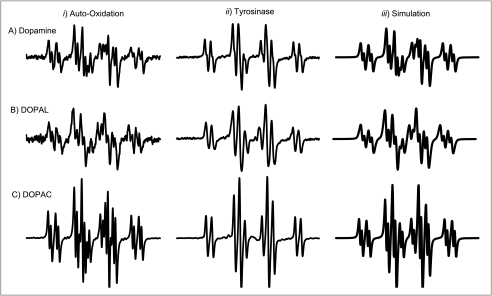
Auto-oxidation (via high pH) of DOPAL also resulted in the formation of a DOPAL semiquinone radical (Fig. 3B, column i). As expected, there are small differences in hyperfine splittings observed in this system, as no divalent cation is present. DA and DOPAC also readily auto-oxidized at pH 10.5 to semiquinone radicals and displayed additional hyperfine splittings (Fig. 3, A and C, column i). Computer simulations reproduced well the experimental spectra (correlation coefficients generally >0.99) (Fig. 3, column iii). DA, DOPAL, and DOPAC oxidized by tyrosinase under spin stabilization conditions (Fig. 3, column ii) are also shown for comparison. The spin-stabilized radicals were also simulated successfully (spectra not shown). Because DOPAL, DOPAC, and DA each has five similarly located hydrogens that can provide hyperfine splittings, they produce similar EPR spectra. The hyperfine splitting constants, reported for the first time for DOPAL, are shown in Table 1. The coupling constants determined for DA and DOPAC are in good agreement with those reported in the literature (6, 36). The g values for DA, DOPAL, and DOPAC are reported in Table 1. Experimental DOPAL radical concentrations were similar to DA for all tested conditions, whereas DOPAC radical levels were severalfold higher (data not shown). Also, incubation of DOPAL (400 μm) with 2.0 eq of H2O2 did not result in the production of a DOPAL radical.
FIGURE 3.
EPR spectra of DA, DOPAL, and DOPAC semiquinone radicals. DA (A), DOPAL (B), and DOPAC (C) radical spectra were generated via high pH-induced auto-oxidation under non-spin-stabilized conditions (column i), Mg2+-stabilized tyrosinase oxidation at pH 7.4 (column ii), and WinSim computer simulation (column iii). The radicals of the dopaminergic compounds are spectrally similar under the same conditions. Also note the hyperfine splitting apparent under conditions of spontaneous oxidation (column i).
TABLE 1.
NMR Spectroscopy
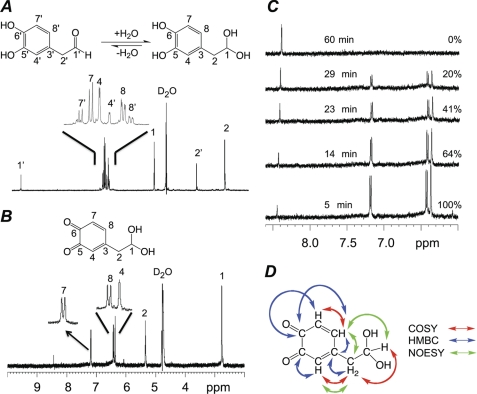
1H NMR spectra of DOPAL in D2O were collected prior to oxidation. Interestingly, these spectra indicated the presence of a well defined mixture of two components. One set of signals matched the expected signals for DOPAL (δ 9.69 (CHO, t, 1H, J = 1.6 Hz), 6.92 (CH aromatic, d, 1H, J = 7.9 Hz), 6.80 (CH aromatic, d, 1H, J = 1.8 Hz), 6.71 (CH aromatic, dd, 1H, J = 7.9, 1.8 Hz), and 3.76 (CH2, d, 1H, J = 1.6 Hz)), whereas the other signals matched the carbonyl hydration product of DOPAL (δ 6.88 (CH aromatic, d, 1H, J = 7.9 Hz), 6.84 (CH aromatic, d, 1H, J = 2.2 Hz), 6.74 (CH aromatic, dd, 1H, J = 7.9, 2.2 Hz), 5.19 (CH(OH)2, t, 1H, J = 5.7), and 2.81 (CH2, d, 1H, J = 5.7 Hz)) (Fig. 4A). Comparative proton integrations indicated that the aldehyde of DOPAL is ∼71% hydrated in aqueous media (Keq values are reported in Table 2). No difference was noted between the hydration levels in D2O versus buffer at pD 7.5.
FIGURE 4.
NMR analysis of the DOPAL quinone. A, 1H NMR of unoxidized DOPAL in aqueous media (D2O) shows an approximate 5:2 ratio of hydrate:aldehyde. Proton assignments are indicated. Aromatic signals are magnified for clarity. B, 1H NMR of DOPAL oxidized by 1.00 eq of NaIO4. As indicated, the resonances fit with the expected structure of the quinone. Aromatic signals are magnified for clarity. C, the stability of the quinone was investigated by measuring the intensity of selected peaks over time (25 °C, D2O). As the quinone disappeared, the hypothesized polymer (8.44 ppm) resonance increased. The percentage of quinone remaining is indicated for each of the spectra. D, NOESY- and two-dimensional NMR-based resonance assignments and NOE correlations verify the ortho-quinone hydrate structure. Correlations useful for structural determination are indicated by colored arrows (red, COSY; blue, HMBC; green, NOESY).
TABLE 2.
Equilibrium constants for DOPAL carbonyl hydration as determined by NMR
Keq values for DOPAL hydrate formation in aqueous media were measured via comparative NMR proton integrations. Values reported are means.
a S.E., standard error of the mean.
b 10 mm NaH2PO4 pD 7.5 in D2O.
Careful oxidation of DOPAL by NaIO4 resulted in the formation of a unique species (Fig. 4B) demonstrating a simplified spectrum and shifted aromatic resonances: δ 7.18 (CH aromatic, d, 1H, J = 10.1 Hz), 6.42 (CH aromatic, d, 1H, J = 10.1 Hz), 6.37 (CH aromatic, s, 1H), 5.34 (CH(OH)2, t, 1H, J = 5.7 Hz), and 2.76 (CH2, d, 1H, J = 5.7 Hz); referenced to acetone-d6 at 2.05 ppm. Under the experimental conditions (D2O, 25 °C) this species was short lived (t½ < 60 min; Fig. 4C). As these peaks diminished, a new single peak at 8.44 ppm increased concurrently, which may have been due to the formation of a polymer; this interpretation is also consistent with the formation of a broad hump under the aromatic resonances during the time course of the reaction. Use of either unbuffered or buffered D2O gave rise to the same oxidized product; however, unbuffered D2O minimized the formation of the 8.44-ppm polymer-like peak, which enhanced the intensities of the other resonances. The special conditions developed (−20 °C, 1:4 dilution with cold acetone) enhanced the lifetime of the species significantly, allowing completion of COSY, NOESY, and other heteronuclear two-dimensional NMR experiments. Importantly, these conditions did not change the course of the reaction or the product formed. Peak assignments and bond correlations apparent from NMR experiments are indicated schematically in Fig. 4D. 13C shifts were determined by HMQC and HMBC experiments (δ 180.95 (CO, aromatic), 180.30 (CO, aromatic), 152.49 (C, aromatic), 144.11 (CH, aromatic), 128.90 (CH, aromatic), 128.80 (CH, aromatic), 89.76 (CH(OH)2), and 44.40 (CH2)). This structure is best described as the ortho-quinone of DOPAL with a hydrated aldehyde. These experiments confirmed for the first time the formation of a DOPAL quinone.
Protein Reactivity
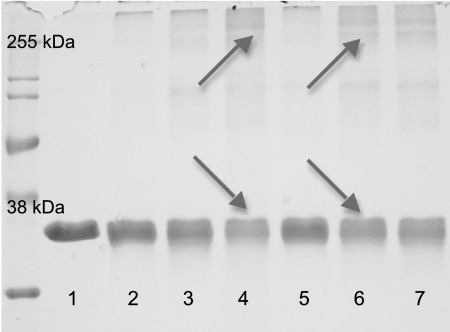
The oxidation of DOPAL enhanced its ability to induce protein cross-linking of the model protein, GAPDH (Fig. 5). The different oxidation conditions represent different active components (NaIO4, 10 s: radical-quinone mixture; NaIO4, 60 s: quinone only; tyrosinase, 10 s: radical-catechol mixture; tyrosinase, 60 s: radical-quinone mixture; K3Fe(CN)6: radical only). Non-oxidized DOPAL caused ∼17% loss of protein as compared with the control (Table 3). Oxidation with NaIO4 at the early and late time points resulted in further losses of 11 and 6%, respectively. Tyrosinase oxidation increased protein losses by 30% when tyrosinase was added at 10 s and 22% when added at 60 s. Unlike NaIO4 and tyrosinase, control reactions of K3Fe(CN)6 with GAPDH (without DOPAL) did cause some loss of protein (∼12%). Therefore, DOPAL oxidized by K3Fe(CN)6 was compared with K3Fe(CN)6-treated protein instead of untreated protein (Fig. 5, lane 1), indicating a 29% greater protein loss. Concurrent with the disappearance of the control band protein is the formation of higher molecular weight aggregates indicative of protein cross-linking (note arrows in Fig. 5). Overall, the oxidation of DOPAL enhanced cross-linking by up to 30%.
FIGURE 5.
Oxidation of DOPAL-enhanced GAPDH cross-linking. SDS-PAGE and densitometry were used to compare the ability of DOPAL oxidized by various methods to induce cross-linking of the model protein, GAPDH. 0.3 mg/ml protein in 50 mm NaH2PO4, pH 7.4, was reacted with 50 μm DOPAL for 4 h at 37 °C. Cross-linking is apparent as a loss of control protein concurrent with the formation of higher molecular weight species (indicated by arrows). Conditions and results for each lane are as noted in Table 3.
TABLE 3.
Conditions and results for Fig. 5
| Lane | Conditions | Reactive species | Resultsa |
|---|---|---|---|
| % | |||
| 1 | Control | NA | NA |
| 2 | DOPAL alone | Catechol | |
| 3 | NaIO4b | Radical-Quinone | 11 |
| 4 | Tyrosinaseb | Catechol-Radical | 30 |
| 5 | NaIO4c | Quinone | 6 |
| 6 | Tyrosinasec | Radical-Quinone | 22 |
| 7 | K3Fe(CN)6 | Radical | 29 |
a Non-oxidized DOPAL (“DOPAL alone”) caused a 17% loss of protein. Results are the amount beyond 17%.
b 10 s pre-oxidation (mimics early oxidation product).
c 60 s pre-oxidation (mimics later oxidation product).
DOPAL Auto-oxidation and ROS Production
DOPAL undergoes relatively slow auto-oxidation to quinone (Fig. 6A). In the presence of SOD (8 μm), the rate of auto-oxidation is accelerated. A lag period precedes the rate increase. Increasing the SOD by a factor of 5 (40 μm) dramatically increased the rate of auto-oxidation and nearly eliminated the lag period. The SOD concentrations used here (8, 40 μm) mimic the levels in the cytoplasm (6–7 μm) and mitochondria (42 μm) (37). These results (i.e. enhancement of the rate of auto-oxidation upon the addition of SOD to the reaction mixture) indicate that air oxidation of DOPAL results in the formation of superoxide (Fig. 6B). Equilibrium for this reaction generally favors reactants (30). However, the presence of SOD can shift the equilibrium toward the products by rapid dismutation of superoxide, effectively “pulling” the reaction.
FIGURE 6.
Enhancement of the rate of DOPAL auto-oxidation by SOD indicates that superoxide is being formed. A, auto-oxidation of DOPAL at pH 7.8 proceeds slowly but is greatly accelerated in the presence of increasing concentrations of SOD. B, although superoxide is formed during spontaneous (i.e. air) oxidation of catechols and hydroquinones, the reverse reaction is much faster. Sufficient removal of superoxide by SOD allows the reaction to proceed much more quickly.
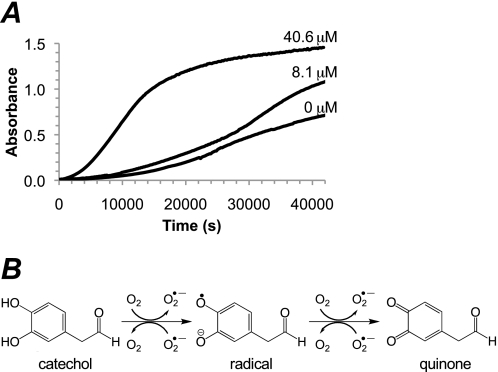
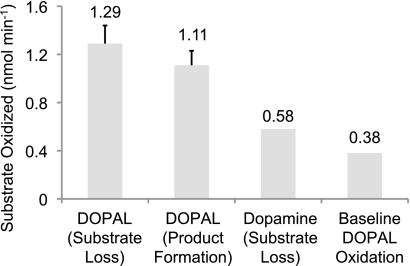
COX-2 Oxidation of DOPAL
The turnover numbers (kcat) for the oxidation of DOPAL and DA by COX-2 were calculated by determining the initial linear slopes (Fig. 7 and Table 4). The results indicate that a small but significant portion of DOPAL is oxidized non-enzymatically by assay components. The reported rates were corrected to account for this base-line oxidation. DOPAL had a kcat of 6.96 ± 0.40 and 5.54 ± 0.89 min−1 as calculated by substrate loss and product formation, respectively; the difference in the values between the two methods arises due to the reactive and unstable nature of radicals and quinones. In comparison, DA had a kcat of 4.18 min−1, which is in good agreement with the literature (38). Furthermore, oxidized DOPAL demonstrated sensitivity to the acidic conditions utilized in the work-up procedure for this assay.
FIGURE 7.
Summary of the initial linear slopes of DOPAL oxidized by COX-2. DOPAL (50 μm) was oxidized by 100 units/ml COX-2. The results, based on the concentration of substrate oxidized, were calculated using both product formation and substrate loss. DA (65 μm) was also shown for comparison using substrate disappearance to calculate the initial linear slope. The assay system without the added enzyme functioned as a control and allowed the results to be corrected for base-line DOPAL oxidation.
TABLE 4.
Summary of COX-2 kcat data
Initial linear slopes were obtained from the oxidation of DOPAL (50 μm) or DA (65 μm) by 100 units/ml COX-2 (100 mm KH2PO4, pH 7.4, 100 μm potassium arachidonate, 1 μm hematin, 1 ml vol). DOPAL kcat values were calculated via both substrate loss and product formation. The DA kcat value was calculated from substrate disappearance only and served as a comparison. Non-enzymatic oxidation of DOPAL served as a control, and the levels of base-line oxidation were subtracted from the noted results.
| Conditions | kcata |
|---|---|
| min−1 | |
| Dopamine (substrate loss) | 4.18 |
| Dopamine (literature value)b | 3.96 |
| DOPAL (substrate loss) | 6.96 ± 0.40 |
| DOPAL (product formation) | 5.54 ± 0.89 |
a kcat is reported ±S.E.
b Mattammal et al. (38).
DISCUSSION
Cytosolic DA can undergo catechol oxidation (2, 39) to form radicals and quinones that exert cytotoxicity through a variety of mechanisms (6, 12). Alternatively, DA may undergo monoamine oxidase-catalyzed oxidative deamination to produce DOPAL, which can be further metabolized by aldehyde dehydrogenase to DOPAC (16, 40). Previously, we showed that lipid peroxidation, which is closely associated with oxidative stress, can inhibit aldehyde dehydrogenase causing elevated intracellular DOPAL levels (17, 24). DOPAL is highly toxic and is hypothesized to be a causative factor in PD (15, 22). Furthermore, catechol oxidation may be important for the toxicity of DOPAL.
Tyrosinase and NaIO4 are known to oxidize DA to a quinone. The oxidation of DOPAL by these methods produced two unique species. EPR confirmed that the initial product was a novel DOPAL semiquinone radical (Scheme 2) produced experimentally at concentrations similar to the DA radical (data not shown). Importantly, the DA semiquinone is a known toxic agent. It interferes with DA storage, oxidatively damages DNA and proteins, produces ROS, and undergoes redox cycling (5, 6).
The DOPAC radical was present in much higher concentrations than either DA or DOPAL, under identical experimental conditions. However, DOPAC has limited toxicity. This is likely because of its further metabolism or selective removal via a transporter (41). Therefore, the DOPAC radical formation reported here may have limited biological consequences.
NMR was used to identify the second oxidation product of DOPAL as the ortho-quinone, allowing us to uncover the carbonyl hydration of DOPAL. Although hydration (gem-diol formation) in aqueous media is known for some aldehydes (42), it has been previously unreported for DOPAL. Importantly, the prevalence of the hydrate over the aldehyde (Keq = 2.50) calls into question the role of the aldehyde versus the gem-diol in the behavior of DOPAL, at both normal and toxic levels.
1H NMR resonances of the later DOPAL oxidation product were consistent with an ortho-quinone structure, which was verified with NOESY, COSY, HMQC, and HMBC experiments. Interestingly, upon oxidation of DOPAL to the quinone, aldehyde resonances were no longer present or were below the detection limit of the NMR. This disappearance is likely because of a shift in the equilibrium for the hydration reaction. Generally, Keq shifts away from the aldehyde are catalyzed by strongly electronegative substituents. For example, the Keq for acetaldehyde hydration is 1.06, but for trifluoroacetaldehyde it is 2.9 × 104 (42). The Keq of 2.50 for DOPAL hydration indicates that the catechol is a strong electron-withdrawing group. However, the apparent loss of the aldehyde resonance following oxidation indicates that the quinone is much more electronegative than the catechol. The Keq of hydration for the DOPAL quinone is likely several orders of magnitude higher than the catechol. These results indicate that the DOPAL quinone is highly electrophilic and may be susceptible to nucleophilic attack and/or reduction.
Because of the similar electrophilic nature and cellular localization, the DA and DOPAL quinones likely have comparable biological effects. Toxicity mechanisms for the DA quinone include nucleophilic reactivity with proteins and thiols, depletion of cellular reducing equivalents, and ROS production (12).
Protein reactivity is an important mechanism of toxicity for both DOPAL and oxidized DA (43). We have shown here that DOPAL oxidation enhances its ability to induce cross-linking of GAPDH. Because of its role as a redox-sensitive enzyme involved in glucose metabolism and multiple external thiol residues (12) and its cellular proximity to DA metabolism, GAPDH serves as an appropriate target for modification by DOPAL. Also, DA-induced aggregation of GAPDH results in increased oxidative stress and cytotoxicity to dopaminergic cell models (44, 45). Although both the DOPAL radical and quinone increased protein cross-linking, this effect was more pronounced for the radical (29%) than for the quinone (6%). This may indicate that GAPDH is more susceptible to radical-induced oxidative damage than quinone-induced nucleophilic reactivity. Although quinones are susceptible to Michael-type ring addition reactions with nucleophiles such as thiols, this mechanism did not seem to contribute greatly to overall protein reactivity. Also, reductive gel conditions indicate that radical-induced cross-linking is due to oxidative damage above and beyond simple disulfide rearrangements. The protein aggregates formed here may involve stable sulfhydryl cross-links (46) or other types of oxidative damage such as peroxidized proteins or protein carbonyls. A high increase (22%) in protein reactivity was evident with the radical-quinone mixture, indicating that redox cycling and ROS production may contribute to the observed results. Furthermore, GAPDH is known to be sensitive to ROS modification, and GAPDH aggregation is involved in dopaminergic models of oxidative stress-induced cell death (45). This makes the cross-linking observed here potentially relevant as a specific mechanism of toxicity for DOPAL.
Another toxicity mechanism for DOPAL may be ROS production. Our results indicate that superoxide is produced during DOPAL auto-oxidation. Superoxide can be formed via sequential one-electron reductions of O2 (Fig. 6B), although the reverse reaction is usually favored (47). The varying lag periods and the biphasic nature of the reaction (Fig. 6A) are consistent with the need to generate enough DOPAL quinone to allow sufficient semiquinone to be formed to bring about the “rapid” oxidation of DOPAL (33, 47). That SOD reduces the lag time is consistent with the formation of superoxide in this system via the semiquinone radical.
Also, DOPAL reportedly will react with H2O2, producing a hydroxyl radical (20). The proposed mechanism involves one-electron oxidation of DOPAL and concurrent reduction of H2O2 to the hydroxyl radical. However, EPR experiments of DOPAL incubated with H2O2 did not result in the formation of a DOPAL radical. Therefore, H2O2 production by SOD was not a significant contributor to DOPAL auto-oxidation in these experiments.
Enhancement of DOPAL auto-oxidation is highly relevant given that SOD is up-regulated in the substantia nigra of PD patients (48, 49). Superoxide dismutation may be beneficial for cells but also result in the deleterious consequence of increased formation of DOPAL radicals and quinones.
COX-2 is a bifunctional, inducible, xenobiotic-metabolizing enzyme (50) capable of oxidizing catechols and catecholamines as cosubstrates during prostaglandin biosynthesis (39). DA is a known COX-2 substrate and can be oxidized to a reactive semiquinone radical or quinone capable of modifying DNA and proteins. Elevated activity and expression of COX-2 occurs in the substantia nigra in PD and may contribute to neurodegeneration via oxidation of biogenic amines to endogenous neurotoxins. Reported sites of COX-2 up-regulation vary conflictingly between dopaminergic neurons, non-astrocytic glial cells, or both (51–53). The COX-2 catalyzed oxidation of DA may also be involved in accumulation of α-synuclein, the primary component of Lewy bodies (39). Furthermore, certain COX-2 inhibitors prevent or attenuate PD in human epidemiological studies and neurotoxin-induced in vitro and animal models. This suggests a pivotal role for COX-2 in the pathogenesis of PD (54–57). Here, we have shown for the first time that DOPAL is also a substrate for COX-2. This development suggests a relevant mechanism of formation for the DOPAL radical and quinone in cells affected by PD. Although Schiff base formation is assumed to be essential for the reactivity of DOPAL with proteins and the resulting cellular dysfunction, catechol oxidation is likely also important for the mediation of DOPAL toxicity. Furthermore, some of the neurodegeneration previously associated with DA oxidation may involve the oxidation of other species such as DOPAL. These findings have implications for the role of DOPAL in PD pathogenesis and therapeutic development.
We have shown here for the first time the oxidation of DOPAL to a semiquinone radical and an ortho-quinone. Such oxidations result in ROS formation and enhanced cross-linking of proteins. The reactivity of oxidized DOPAL with GAPDH serves as an example of a potentially deleterious biological outcome. Also, DOPAL was demonstrated to be a COX-2 substrate, indicating a method for the possible biological formation of the oxidized products identified here. The findings of this work highlight the potential importance of DOPAL oxidation by COX-2 (or other methods) in PD.
Supplementary Material
Acknowledgments
We gratefully acknowledge the technical assistance of Brett A. Wagner and T. Joost van't Erve with EPR spectroscopy.
This work was supported, in whole or in part, by National Institutes of Health Grants R01ES15507 (to J. A. D.), R01GM073929 and P42ES013661 (to G. R. B.), and T32GM 008365 from NIGMS (to D. G. A.). This work was also supported by the University of Iowa College of Pharmacy and a fellowship through the American Foundation for Pharmaceutical Education (to D. G. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6 and Tables 1 and 2.
- PD
- Parkinson disease
- COSY
- 1H-1H correlated spectroscopy
- DA
- dopamine
- DMSO-d6
- perdeuterated dimethyl sulfoxide
- DOPAL
- 3,4-dihydroxyphenylacetaldehyde
- DOPAC
- 3,4-dihydroxyphenylacetic acid
- HMBC
- 1H-13C heteronuclear multiple bond correlation
- HMQC
- 1H-13C heteronuclear multiple quantum correlation
- NOESY
- 1H-1H nuclear Overhauser effect spectroscopy
- SOD
- superoxide dismutase
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Davie C. A. (2008) Br. Med. Bull. 86, 109–127 [DOI] [PubMed] [Google Scholar]
- 2. Chinta S. J., Andersen J. K. (2008) Biochim. Biophys. Acta 1780, 1362–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fornstedt B., Brun A., Rosengren E., Carlsson A. (1989) J. Neural Transm. Park. Dis. Dement. Sect. 1, 279–295 [DOI] [PubMed] [Google Scholar]
- 4. Collins M. A., Neafsey E. J. (2002) Neurotoxicol. Teratol. 24, 571–577 [DOI] [PubMed] [Google Scholar]
- 5. Terland O., Almas B., Flatmark T., Andersson K. K., Sorlie M. (2006) Free Radic. Biol. Med. 41, 1266–1271 [DOI] [PubMed] [Google Scholar]
- 6. Goncalves L. L., Ramkissoon A., Wells P. G. (2009) Chem. Res. Toxicol. 22, 842–852 [DOI] [PubMed] [Google Scholar]
- 7. Graham D. G. (1978) Mol. Pharmacol. 14, 633–643 [PubMed] [Google Scholar]
- 8. Graham D. G., Tiffany S. M., Bell W. R., Jr., Gutknecht W. F. (1978) Mol. Pharmacol. 14, 644–653 [PubMed] [Google Scholar]
- 9. Stokes A. H., Hastings T. G., Vrana K. E. (1999) J. Neurosci. Res. 55, 659–665 [DOI] [PubMed] [Google Scholar]
- 10. Halliwell B. (1992) J. Neurochem. 59, 1609–1623 [DOI] [PubMed] [Google Scholar]
- 11. Hastings T. G., Lewis D. A., Zigmond M. J. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 1956–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Akagawa M., Ishii Y., Ishii T., Shibata T., Yotsu-Yamashita M., Suyama K., Uchida K. (2006) Biochemistry 45, 15120–15128 [DOI] [PubMed] [Google Scholar]
- 13. Burke W. J., Li S. W., Chung H. D., Ruggiero D. A., Kristal B. S., Johnson E. M., Lampe P., Kumar V. B., Franko M., Williams E. A., Zahm D. S. (2004) Neurotoxicology 25, 101–115 [DOI] [PubMed] [Google Scholar]
- 14. Burke W. (2003) Curr. Drug Targets CNS Neurol. Disord. 2, 143–148 [DOI] [PubMed] [Google Scholar]
- 15. Burke W. J., Li S. W., Williams E. A., Nonneman R., Zahm D. S. (2003) Brain Res. 989, 205–213 [DOI] [PubMed] [Google Scholar]
- 16. Marchitti S. A., Deitrich R. A., Vasiliou V. (2007) Pharmacol. Rev. 59, 125–150 [DOI] [PubMed] [Google Scholar]
- 17. Rees J. N., Florang V. R., Anderson D. G., Doorn J. A. (2007) Chem. Res. Toxicol. 20, 1536–1542 [DOI] [PubMed] [Google Scholar]
- 18. Rees J. N., Florang V. R., Eckert L. L., Doorn J. A. (2009) Chem. Res. Toxicol. 22, 1256–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kristal B. S., Conway A. D., Brown A. M., Jain J. C., Ulluci P. A., Li S. W., Burke W. J. (2001) Free Radic. Biol. Med. 30, 924–931 [DOI] [PubMed] [Google Scholar]
- 20. Li S. W., Lin T. S., Minteer S., Burke W. J. (2001) Mol. Brain Res. 93, 1–7 [DOI] [PubMed] [Google Scholar]
- 21. Hashimoto T., Yabe-Nishimura C. (2002) Brain Res. 931, 96–99 [DOI] [PubMed] [Google Scholar]
- 22. Mattammal M. B., Haring J. H., Chung H. D., Raghu G., Strong R. (1995) Neurodegeneration 4, 271–281 [DOI] [PubMed] [Google Scholar]
- 23. Fellman J. H. (1958) Nature 182, 311–312 [DOI] [PubMed] [Google Scholar]
- 24. Jinsmaa Y., Florang V. R., Rees J. N., Anderson D. G., Strack S., Doorn J. A. (2009) Chem. Res. Toxicol. 22, 835–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li S. W., Spaziano V. T., Burke W. J. (1998) Bioorg. Chem. 26, 45–50 [Google Scholar]
- 26. Tsukamoto S., Kato H., Hirota H., Fusetani N. (1994) Tetrahedron 50, 13583–13592 [Google Scholar]
- 27. Buettner G. R. (1986) Free Radic. Res. Commun. 1, 349–353 [DOI] [PubMed] [Google Scholar]
- 28. Kalyanaraman B., Sealy R. C. (1982) Biochem. Biophys. Res. Commun. 106, 1119–1125 [DOI] [PubMed] [Google Scholar]
- 29. Kalyanaraman B., Felix C. C., Sealy R. C. (1985) Environ. Health Perspect. 64, 185–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Song Y., Wagner B. A., Lehmler H. J., Buettner G. R. (2008) Chem. Res. Toxicol. 21, 1359–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Duling D. R. (1994) J. Magn. Reson. B 104, 105–110 [DOI] [PubMed] [Google Scholar]
- 32. Adams M., Blois M. S., Sands R. H. (1958) J. Chem. Phys. 28, 774–776 [Google Scholar]
- 33. Eyer P. (1991) Chem. Biol. Interact. 80, 159–176 [DOI] [PubMed] [Google Scholar]
- 34. Wangpradit O., Mariappan S. V., Teesch L. M., Duffel M. W., Norstrom K., Robertson L. W., Luthe G. (2009) Chem. Res. Toxicol. 22, 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Land E. J., Simic M., Swallow A. J. (1971) Biochim. Biophys. Acta 226, 239–240 [DOI] [PubMed] [Google Scholar]
- 36. Kirima K., Tsuchiya K., Yoshizumi M., Kameda T., Houchi H., Azuma M., Tamaki T. (2001) Chem. Pharm. Bull. (Tokyo) 49, 576–580 [DOI] [PubMed] [Google Scholar]
- 37. Chang L. Y., Slot J. W., Geuze H. J., Crapo J. D. (1988) J. Cell Biol. 107, 2169–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mattammal M. B., Strong R., Lakshmi V. M., Chung H. D., Stephenson A. H. (1995) J. Neurochem. 64, 1645–1654 [DOI] [PubMed] [Google Scholar]
- 39. Chae S. W., Kang B. Y., Hwang O., Choi H. J. (2008) Neurosci. Lett. 436, 205–209 [DOI] [PubMed] [Google Scholar]
- 40. Eisenhofer G., Kopin I. J., Goldstein D. S. (2004) Pharmacol. Rev. 56, 331–349 [DOI] [PubMed] [Google Scholar]
- 41. Lamensdorf I., He L., Nechushtan A., Harvey-White J., Eisenhofer G., Milan R., Rojas E., Kopin I. J. (2000) Eur. J. Pharmacol. 388, 147–154 [DOI] [PubMed] [Google Scholar]
- 42. Carey F. A., Sundberg R. J. (2000) Advanced Organic Chemistry, Part A: Structure and Mechanisms, 4th Ed., pp. 449–451, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- 43. LaVoie M. J., Ostaszewski B. L., Weihofen A., Schlossmacher M. G., Selkoe D. J. (2005) Nat. Med. 11, 1214–1221 [DOI] [PubMed] [Google Scholar]
- 44. Nakajima H., Amano W., Fukuhara A., Kubo T., Misaki S., Azuma Y. T., Inui T., Takeuchi T. (2009) Biochem. Biophys. Res. Commun. 390, 1066–1071 [DOI] [PubMed] [Google Scholar]
- 45. Nakajima H., Amano W., Kubo T., Fukuhara A., Ihara H., Azuma Y. T., Tajima H., Inui T., Sawa A., Takeuchi T. (2009) J. Biol. Chem. 284, 34331–34341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nakajima H., Amano W., Fujita A., Fukuhara A., Azuma Y. T., Hata F., Inui T., Takeuchi T. (2007) J. Biol. Chem. 282, 26562–26574 [DOI] [PubMed] [Google Scholar]
- 47. Song Y., Buettner G. R. (2010) Free Radic. Biol. Med. 49, 919–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Saggu H., Cooksey J., Dexter D., Wells F. R., Lees A., Jenner P., Marsden C. D. (1989) J. Neurochem. 53, 692–697 [DOI] [PubMed] [Google Scholar]
- 49. Baillet A., Chanteperdrix V., Trocme C., Casez P., Garrel C., Besson G. (2010) Neurochem. Res. 35, 1530–1537 [DOI] [PubMed] [Google Scholar]
- 50. Wangpradit O., Moman E., Nolan K. B., Buettner G. R., Robertson L. W., Luthe G. (2010) Chemosphere 81, 1501–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bartels A. L., Leenders K. L. (2010) Curr. Neuropharmacol. 8, 62–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Minghetti L. (2004) J. Neuropathol. Exp. Neurol. 63, 901–910 [DOI] [PubMed] [Google Scholar]
- 53. Teismann P., Vila M., Choi D. K., Tieu K., Wu D. C., Jackson-Lewis V., Przedborski S. (2003) Ann. N.Y. Acad. Sci. 991, 272–277 [DOI] [PubMed] [Google Scholar]
- 54. Asanuma M., Miyazaki I., Ogawa N. (2004) Curr. Pharm. Des. 10, 695–700 [DOI] [PubMed] [Google Scholar]
- 55. Hunter R. L., Dragicevic N., Seifert K., Choi D. Y., Liu M., Kim H. C., Cass W. A., Sullivan P. G., Bing G. (2007) J. Neurochem. 100, 1375–1386 [DOI] [PubMed] [Google Scholar]
- 56. Vijitruth R., Liu M., Choi D. Y., Nguyen X. V., Hunter R. L., Bing G. (2006) J. Neuroinflammation 3, 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Feng Z., Li D., Fung P. C., Pei Z., Ramsden D. B., Ho S. L. (2003) Neuroreport 14, 1927–1929 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.