Figure 2.
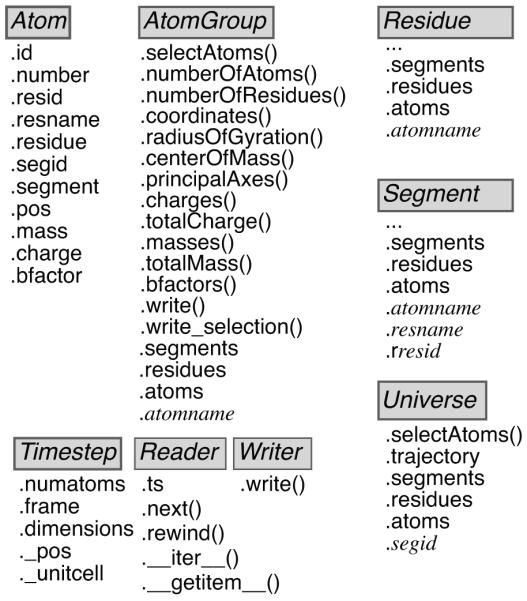
Layout of important MDAnalysis classes. The Universe class contains an AtomGroup of all Atom instances that can be accessed through the attribute Universe.atoms. It also features a number of auto-generated AtomGroups, one for each segment identifier of the topology. Users can obtain information from the individual Atom instances and computed data from a whole AtomGroup by querying the instance attributes and methods. The Timestep class represents the current state of the system at a particular time frame of the trajectory. Trajectory Reader and Writer classes provide an abstract interface to trajectory I/O. They implement special methods that allow “Pythonic” access to the objects such as treating a Reader as an iterator over trajectory frames (via the __iter__() special method) or selecting individual frames by the indexing operation (implemented through a custom __getitem__() method).
