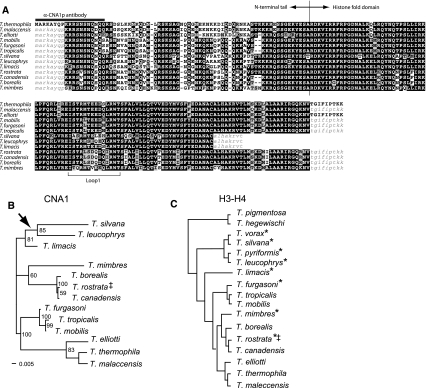
Fig. 2.
Molecular evolution of CNA1 (a) Amino acid alignment of Cna1p from a panel of Tetrahymena species. Highlighted is the border between the N-terminal tail and histone fold domain, the sequence used as an epitope for antibody production, and the highly variable loop 1 region. Sequences corresponding to primer-binding sites are indicated in lower case. b Maximum likelihood phylogeny of CNA1 nucleic acid sequences. Bootstrap values above 50 are indicated at corresponding branches and the scale of substitutions/site is shown. The arrowhead indicates where the tree would root in the context of more diverse species as shown in c. c A phylogenetic tree of Tetrahymena species based on the histone H3-H4 spacer region (adapted from Brunk et al. 1990). The asterisks (*) denote species that lack a germline micronucleus. ‡The identification of this strain as T. rostrata has been challenged (Segade et al. 2009)