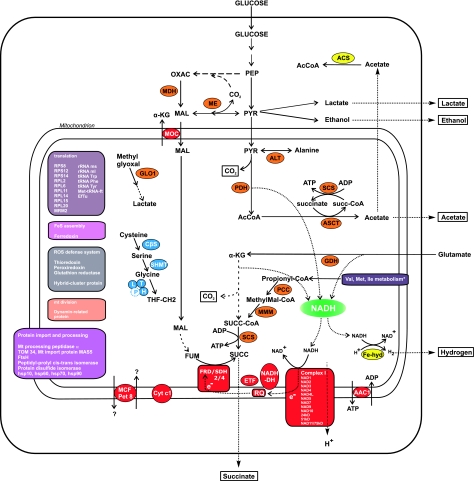
FIG. 3.
Tentative reconstruction of the metabolism of the hydrogen-producing mitochondria of Nyctotherus ovalis. The metabolism is based on proteins that are orthologous to mitochondrial proteins (Methods) and proteins derived from HGT that are likely to have an organellar location based on their metabolic function. It includes the results of metabolic experiments described in Boxma et al. (2005). In red: metabolism linked to NADH (green) oxidation and electron transfer as well as solute carriers. In yellow are the proteins that seem to have been acquired by HGT. In blue: glycine metabolism. In orange: intermediate metabolism. Dotted arrows stand for metabolic steps we assume to be present based on biological experiments, broken arrows for the inferred metabolism for which we did not find the gene yet. AAC: ADP/ATP carrier; ACS: acetyl-CoA synthetase, AMP-forming (EC: 6.2.1.1); ALT: alanine amino transferase; ASCT: acetate:succinate CoA transferase; Cyt c1: cytochrome c1; CβS: cystathione β synthase; EfTu: elongation factor Tu; ETF: electron transfer flavoprotein; Fe-hyd: FeFe hydrogenase; FRD/SDH: fumarate reductase/succinate dehydrogenase; GDH: glutamate dehydrogenase; GLO1: glyoxalase I; MCF Pet 8: mitochondrial carrier family; MDH: malate dehydrogenase; ME: malic enzyme; Met tRNA ft: methionyl-tRNA formyltransferase; MMM: methyl malonyl CoA mutase; MOC: malate:oxoglutarate carrier; NADH-DH: NADH :quinone oxidoreductase; PCC: propionyl CoA carboxylase; PDH: pyruvate dehydrogenase; RQ: rhodoquinone; SCS: succinyl-CoA synthetase; SHMT: serine hydroxymethyl transferase; *includes several enzymes involved in branched-chain amino acid metabolism.