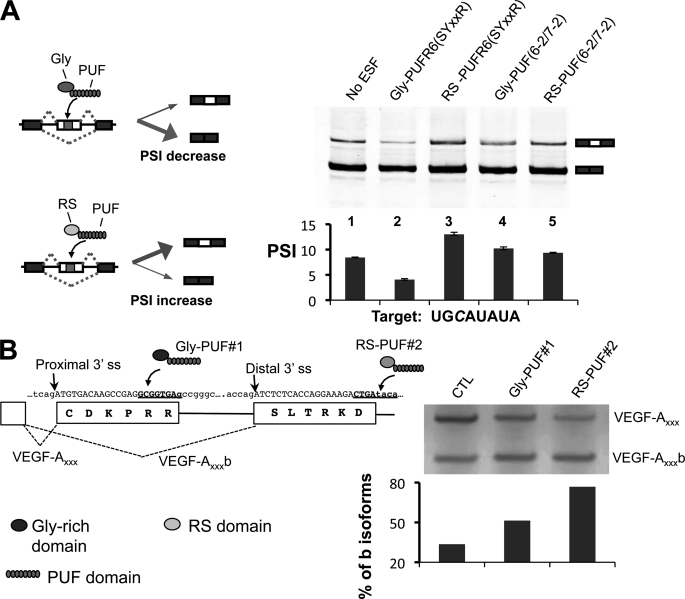
FIGURE 5.
Using the cytosine-recognition code to direct engineered splicing factors. A, modulating alternative splicing of a cassette exon in a reporter RNA. Left, diagram of how the two types of ESFs can affect splicing of a cassette exon. Gly-PUF ESF directed to the exonic target can increase exon inclusion, whereas the RS-PUF ESF can decrease exon inclusion. Top right, RT-PCR products of splicing reactions; bottom right, quantification of splicing. The splicing reporter gene and expression vectors for different ESFs were co-transfected at 1:2 ratio into 293T cells. Total RNA was purified 24 h after transfection, and splicing of the test exon was detected with RT-PCR. The percentage of exon included isoform among all isoforms is represented with PSI value (percentage spliced in). The transfections were carried out in duplicate, and the means of the PSI value were plotted with the error bars indicating the data range. Significant changes (p values are 0.04 and 0.01 for lanes 2 and 3 as judged by paired Student's t test) were observed for ESFs that recognize cognate C-containing target. B, design of ESFs to target endogenous VEGF-A pre-mRNA splicing. The gene and protein sequences of VEGF-A in the region near the alternative splice sites are shown with two PUFs recognizing different cytosine-containing sequences (left panel, underlined sequences). To shift the splicing toward anti-angiogenic VEGF-A isoforms, the cultured MDA-MB-231 cells were transfected with 1 μg of expression vectors of Gly-PUF#1 or RS-PUF#2. Total RNA was purified 24 h after transfection to detect VEGF-A splicing by RT-PCR. The percentages of b isoforms were quantified and are plotted below the gel (left). CTL, control.