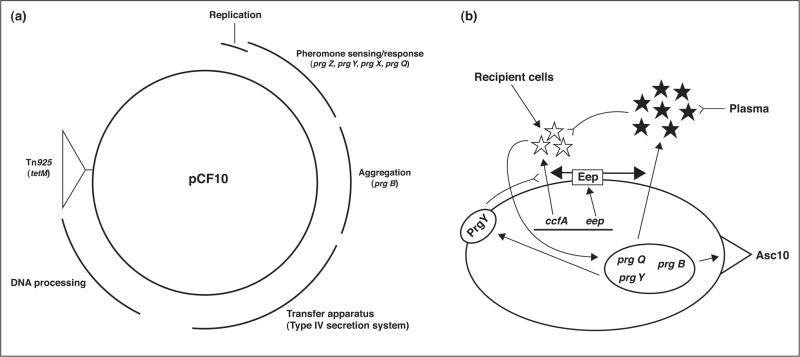
Figure 1.
Genetic organization of pCF10 and the role of extracellular peptide signals in the control of pCF10 conjugation. (a) Map of pCF10, based on DNA sequencing [16] and genetic studies [46]. The curved lines outside the circle denote the segments of the plasmid encoding various biological functions as indicated, with specific genes discussed in the text listed below the respective regions in which they are located. (b) The induction state of pCF10-containing cells is determined by the molar ratios of cCF10 pheromone (white stars; encoded by the chromosomal ccfA gene) to iCF10 inhibitor (black stars; encoded by the prgQ gene of pCF10). Both peptides are processed to their mature heptapeptide form from precursor polypeptides in the membrane by the Eep protease (rectangle in cell membrane). The pCF10-encoded PrgY protein (oval in the membrane) reduces the level of cCF10 produced by its host cell; residual cCF10 escaping PrgY is neutralized by iCF10. Both peptides function by internalization and interaction with PrgX, as described in Figure 2 and in the text, with cCF10 causing increased expression of the prgQ operon leading to production of Asc10, and other proteins involved in conjugation. The peptide ratio can be shifted by recipient cells in close proximity, or in the bloodstream, where iCF10 is sequestered, resulting in self-induction by exogenously produced cCF10. Single arrows indicate a positive effect of one component of the system on another; double-stemmed arrows indicate the polypeptide product of a gene encoded by pCF10 (oval in the cell) or the chromosome (line in the cell); inverted arrows indicate inhibition or destruction of one component by another.