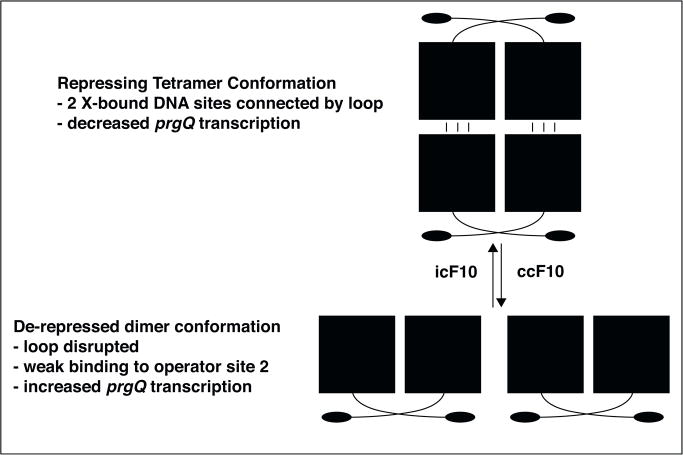
Figure 2.
Pheromone and inhibitor peptides affect PrgX repression of prgQ transcription by altering the oligomerization state of PrgX in pCF10-containing cells. PrgX monomers contain an N-terminal DNA binding domain (filled oval) and a C-terminal regulatory domain (filled square) connected by a short linker region (curved line). Domain swapping promotes formation of a stable dimer that can occupy one operator binding site on pCF10; the region upstream from prgQ contains two such sites. Interaction of iCF10 with PrgX stabilizes a small-C-terminal loop proposed to function as an interface for protein/protein interactions between pairs of dimers to stabilize a tetramer structure (upper panel) where each dimer occupies one operator site on pCF10, and the two sites are connected by a DNA loop (not shown). Pheromone binding to the same region of PrgX causes a structural change at the extreme C-terminus of PrgX that abolishes the tetramer-stabilizing interaction (lower panel), and indirectly reduces operator site occupancy. This increases transcription initiation at prgQ by RNA polymerase.