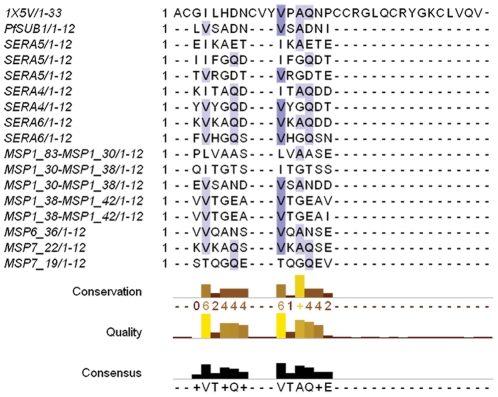
Figure 1. Alignment of PcFK1 with substrate sequences of PfSUB1.
The sequence alignment of PcFK1 (pdb: 1X5V) with the sequences recognized by PfSUB1 shows a comparable residue profile for Site 1 and Site 2. In particular, Site 2 shows the highest sequence similarity with the most conserved residues among the substrate sequences. The shades of blue indicate the degree of conservation among the sequences. The SERA4 and SERA6 processing sites are predicted from sequence alignments and homology with the experimentally determined SERA5 processing sites [4]. All other sites shown here were experimentally determined by amino-acid sequence analysis [5].