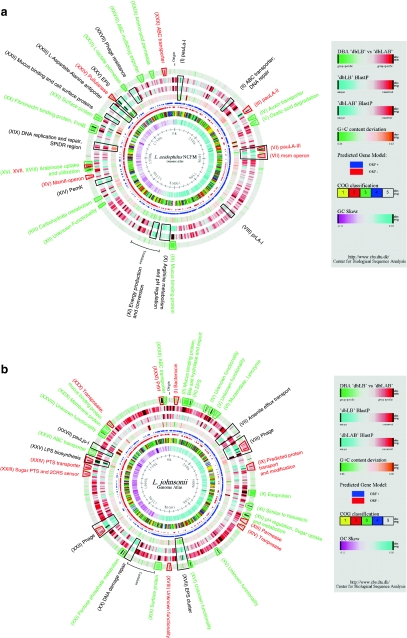
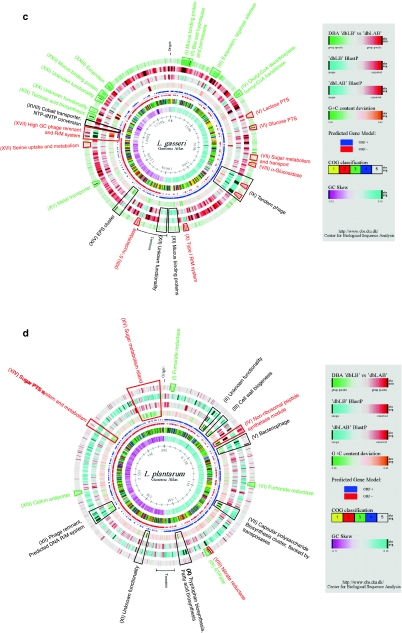
Fig. 1.
Comparative DBA analysis of four Lactobacillus genomes. The atlases each represent a circular view of the complete genome sequences of L. acidophilus NCFM (a), L. johnsonii NCC533 (b), L. gasseri ATCC33323 (c), and L. plantarum WCFS1 (d), respectively. The right-hand legend describes the single circles in the top-down- outermost-innermost direction. The circle was created using Genewiz and in-house developed software. Innermost circle 1, GC-Skew. Circle 2, COG classification. Predicted ORFs were analyzed using the COG database and grouped into the 4 major categories. 1, Information storage and processing; 2, Cellular processes and signaling; 3, Metabolism; 4, Poorly characterized; and 5, ORFs with uncharacterized COGs or no COG assignment. Circle 3, ORF orientation. ORFs in sense orientation (ORF +) are shown in blue; ORFs oriented in anti-sense direction (ORF −) in red. Circle 4, G+C content deviation. Deviations from the average GC content are shown in either green (low GC spike) or orange (high GC spike). A boxfilter was applied to visualize contiguous regions of low or high deviations. Circles 5 and 6, Blast similarities, using LAB and Lactobacillus custom databases, respectively. Deduced amino acid sequences compared against the respective database using gapped BlastP [7]. Regions in blue represent unique proteins in NCFM, whereas highly conserved features are shown in red. The degree of color saturation corresponds to the level of similarity. Circle 7, DBA analysis. Predicted ORFs present in at least one member of the Lactobacillus database (dbLB), but not equally conserved in any member of the LAB database (dbLAB) are highlighted in green. Geneproducts present in at least one member of the LAB database, but not equally conserved in any member of the Lactobacillus database are highlighted in red. Features described in more detail are indicated by trapezoids in the corresponded color and roman numbers. Black trapezoids represent unique genome regions, either not found in any custom database or otherwise outstanding within the genome (color figure online)