FIGURE 1. Tree diagram constructed from the alignment of 554 sequences derived from the HTH_5 family of the Pfam data base.
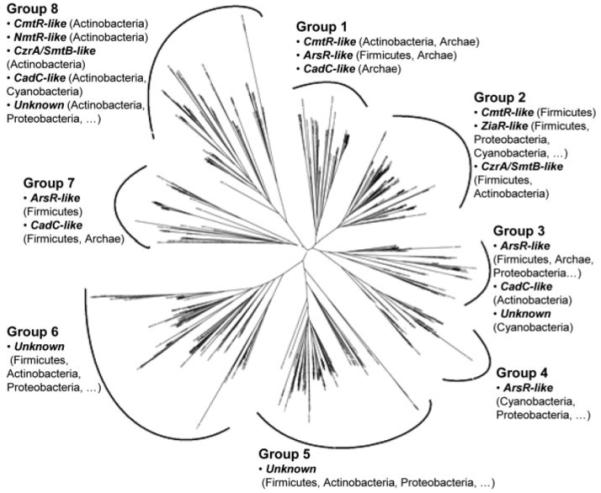
The tree diagram shows eight major groups. The main metal binding motifs occurring in proteins within each group are indicated: CmtR-like sequences possess the CXXXC metal-binding pattern in the α4 helix and potential ligands in the carboxyl-terminal region; CzrA/SmtB-like and NmtR-like possess the pattern DXHX(10)HXX(E/H) in the α5 helix, with the latter also possessing carboxyl-terminal ligands; ArsR-like possess either one or more of (i) CXCXXC, (ii) CXC, or (iii) CXXD in the α3 helix; CadC-like and ZiaR-like possess either one or more of i, ii, and iii in addition to potential ligands in the amino-terminal region, with ZiaR-like also possessing the DXHX(10)HXX(E/H) pattern in α5; the unknown lack these patterns. For each motif, the main bacterial phyla are shown in parentheses (a series of periods indicates sequences are also present from other phyla), and sequences with the motif may be from more than one branch (sub-group).
