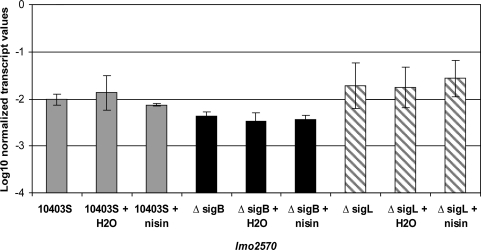
FIG. 1.
Normalized log-transformed lmo2570 transcript levels for Listeria monocytogenes 10403S (gray bars), ΔsigB (black bars), and ΔsigL (hatched bars). Transcript levels were determined by quantitative reverse-transcriptase–polymerase chain reaction using RNA isolated from logarithmic phase (OD600 = 0.4) L. monocytogenes that had been (i) incubated for 10 min after addition of nisin in sterile distilled water to yield a final concentration of 75 AU/mL nisin; (ii) incubated for 10 min after addition of an equivalent volume of sterile distilled water without nisin; or (iii) incubated for 10 min without any addition. Transcript levels were log transformed and normalized to the geometric mean of the transcript levels for the housekeeping genes rpoB and gap. Values represent mean transcript levels from three independent RNA collections; error bars indicate one standard deviation from each mean. Overall ANOVA showed a significant effect of the factor strain, but no effect of the factor condition (i.e., no addition, addition of water, or addition of nisin). Tukey's test showed significantly lower transcript levels for lmo2570 in the ΔsigB strain as compared to the parent strain; transcript levels did not differ significantly between the ΔsigL strain and the parent strain.