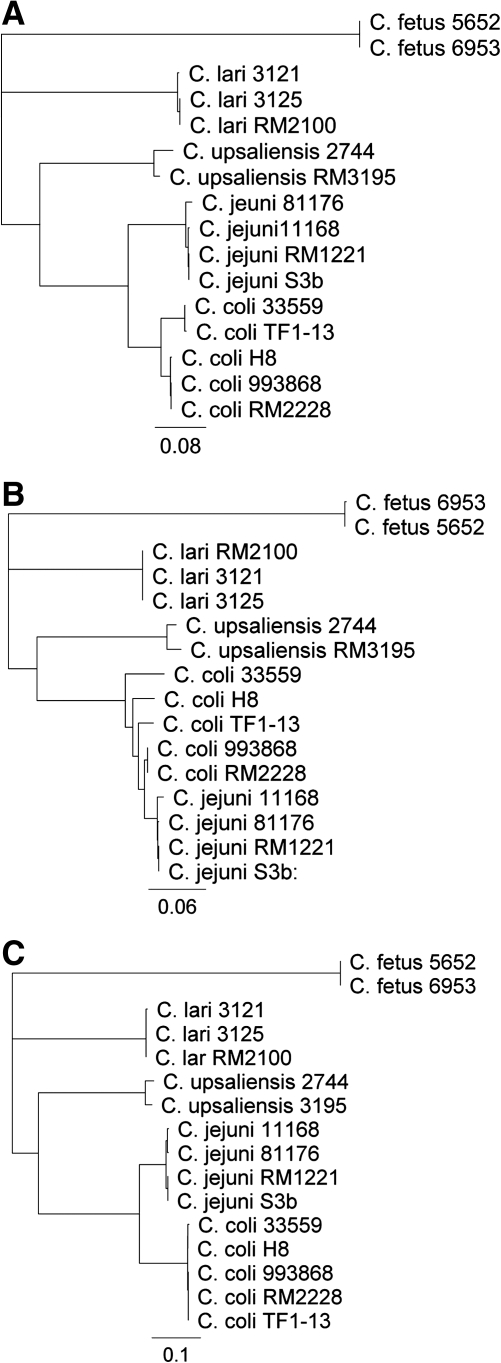
FIG. 1.
Dendrograms constructed with the amino acid sequences of CmeA (A), CmeB (B), and CmeC (C) from different Campylobacter species. The rooted trees were constructed with the neighbor-joining method based on ClustalW alignments of the CmeA, CmeB, and CmeC amino sequences of Camplylobacter strains from this study (C. jejuni S3b; C. coli 993868, 33559, TF1-13, and H-8; C. upsaliensis 2744; C. lari 3121and 3125; C. fetus 5652 and 6953) and representative genome sequences deposited in GenBank (C. jejuni 11168, 81176, and RM1221; C. coli RM2228; C. upsaliensis RM3195; and C. lari RM2100).