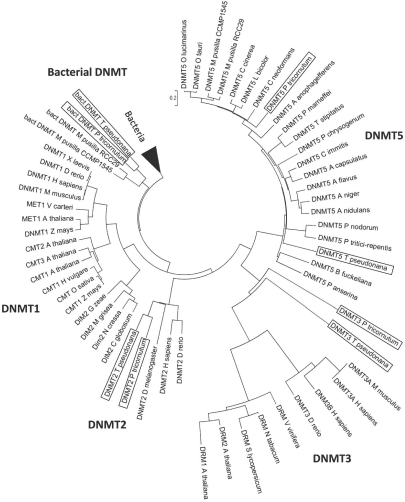
Fig. (4).
DNMTs phylogenetic reconstruction. Bootstrap-supported phylogenetic tree constructed with the Neighbor-Joining method (1000 iterations) showing the relationships between the DNA methyltransferase domains of selected DNMT families. Bacterial DNMTs, indicated by a black dashed curve, come from the following species: Bacillus subtilis, Clostridium cellulolyticum H10, Clostridium kluyveri DSM 555, Moraxella sp., Legionella pneumophila str. Paris, Legionella pneumophila str. Lens, Shewanella sp. MR-4, Roseobacter litoralis Och 149, Neisseria meningitidis MC58, Sulfurimonas denitrificans DSM 1251, Bacteroides fragilis YCH46, Microcystis aeruginosa PCC 7806, Microcystis aeruginosa NIES-843, Dactylococcopsis salina, Cyanothece sp. ATCC 51142, Collinsella stercoris DSM 13279, and Mycoplasma arthritidis 158L3-1. Fungal DNMT5 proteins, indicated by a grey dashed curve, come from the following species: Ajellomyces capsulatus NAm1, Aspergillus flavus NRRL3357, Aspergillus nidulans FGSC A4, Aspergillus niger, Botryotinia fuckeliana B05.10, Coccidioides immitis RS, Coprinopsis cinerea okayama7 (#130), Cryptococcus neoformans var. neoformans B3501A, Laccaria bicolor S238NH82, Penicillium chrysogenum Wisconsin 541255, Penicillium marneffei ATCC 18224, Phaeosphaeria nodorum SN15, Podospora anserina, Pyrenophora triticirepentis Pt1CBFP, and Talaromyces stipitatus ATCC 10500. The tree was drawn using the SplitsTree4 software [107].