Figure 5.
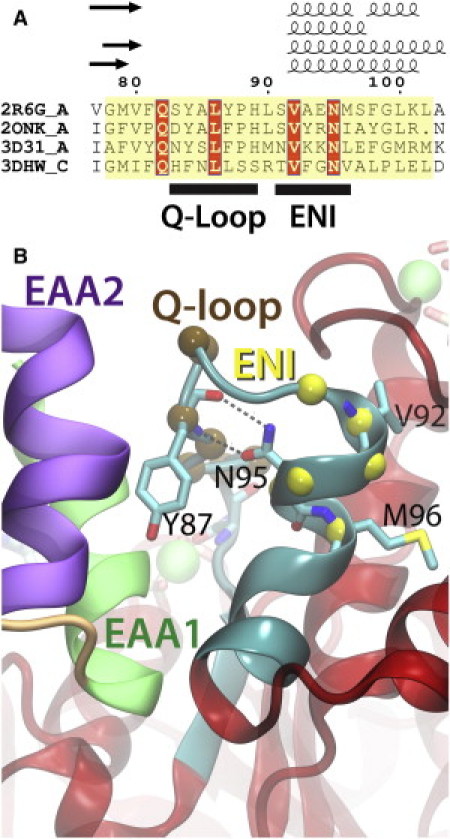
TMD-coupling motifs in MalK. (A) Structure-based sequence alignment of the NBDs in small ABC importers at the NBD-TMD coupling region. The NBD of each crystal structure is structurally aligned to reach the best fit up to the position of G235 of MalK, neglecting the attached regulatory domains or the associated dimerizing helices. The structure-based alignment is performed using Multiseq (40), manually optimized at the Walker A motifs (due to structural variations resulted from nucleotide binding), and formatted with ESPript (41). The full alignment is shown in Fig. S1. (B) A close-up of the Q-loop and ENI motifs in MalK. Structures are colored as in Fig. 4E, and key residues involving the functional role of the ENI motif (Y87, V92, N95, and M96) are shown in stick models. The hydrogen bonds connecting the side chain of N95 and the backbone of Y87 are highlighted.
