Figure 1.
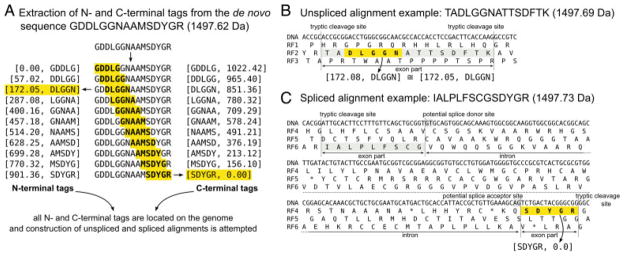
GPF peptide alignment procedure. (A) From the de novo predicted amino acid sequence GDDLGGNAAMSDYGR, all possible amino acid pentamers are extracted with both their N- and C-terminal tryptic fragment masses. Each of these pentamer/fragment mass pairs is located within the genomic DNA sequence with the help of a previously compiled index. From the determined locus, the deduction of unspliced and spliced alignments is attempted. (B) From the N-terminal tag [172.05, DLGGN], a matching peptide can be constructed by N- and C-terminal extension toward the adjoining tryptic cleavage sites. The mass of the resulting peptide TADLGGNATTSDFTK is sufficiently similar for the peptide to be considered a valid GPF peptide candidate. (C) For the C-terminal tag [SDYGR, 0.0], an unspliced alignment cannot be deduced because N-terminal extension of the tag would result in the peptide QSDYGR, which is too light to match the target mass. Deduction of spliced alignments is attempted by searching for all dinucleotide pairs of GT and AG, a common splice donor/acceptor motif in C. reinhardtii, within a user-defined maximum intron length. In this example, the spliced peptide IALPLFSCGSDYGR is deduced and added to the list of GPF peptide candidates. Note: In the right hand side of the figure, an amino acid inferred from a nucleotide triplet is shown centered to the three corresponding nucleotides.