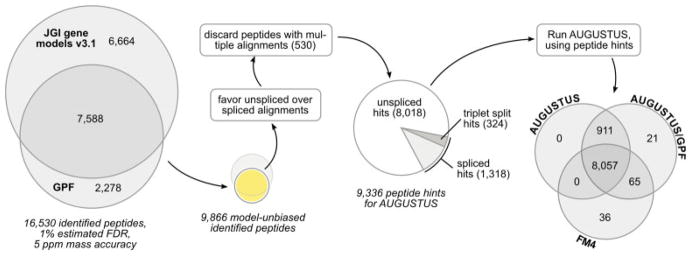
Figure 3.
Processing of OMSSA results. The OMSSA results show that 53% of the identified gene model peptides can be independently confirmed via PEAKS/GPF. Additionally, a set of 2278 peptides identified via PEAKS/GPF alone is available. The set of peptides identified via PEAKS/GPF is used to provide a list of peptide hints for AUGUSTUS. The following filtering steps are applied to this set: (i) whenever an unspliced alignment is available for a peptide, all spliced alignments of the same peptide are discarded because the unspliced variant is the most probable explanation. (ii) Peptides with multiple alignments on the genomic DNA are discarded in order to provide AUGUSTUS with unambiguous peptide hints. A comparison of the resulting AUGUSTUS gene sets with and without the GPF peptide hints to the FM4 gene set reveals an increased amount of coding sequences in the AUGUSTUS gene sets. The AUGUSTUS/GPF gene set confirms most peptides already present in the AUGUSTUS gene set without the peptide hints but introduces 21 new peptides. In addition, 65 peptides that were present in FM4 but not in AUGUSTUS are retained in the AUGUSTUS/GPF gene set.