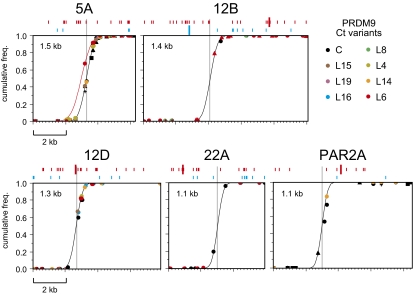
Fig. 3.
Sperm cross-over distributions across genomic intervals regulated by PRDM9 Ct variants. Cross-over molecules from men carrying the Ct allele indicated at top right were mapped by using internal SNPs. Different men carrying the same Ct allele are indicated by different symbols of the same color. Cumulative cross-over frequencies for each man were estimated by mapping 25–215 cross-overs in each orientation and combining reciprocal cross-over data; the only exceptions were at hotspot PAR2A, where two C carriers and the L14 carrier were mapped in only one orientation. Data from all men were combined to estimate the least-squares best-fit cumulative normal distribution (black line) at each hotspot (3). The L6 carrier at hotspot 5A shows a cross-over distribution (red line) significantly shifted by 0.3 kb relative to the hotspot in other tested men (Fisher exact test on numbers of cross-overs mapping 5′ and 3′ to the central C/T SNP rs13355978 in the L6 carrier vs. three other mapped C/T heterozygotes, P < 10−6). The best-fit distributions were used to estimate the center of each hotspot (gray line) and its width within which 95% of cross-overs are located, as indicated. Sequence matches with the binding motif predicted for PRDM9 Ct variants (CCNCNNTNNNCNTNNC) (2) and the A variant (CCNCCNTNNCCNC) (4, 6) are shown above each graph in red and blue, respectively, with perfect matches indicated by large lines and single base mismatches by small lines. Coordinates of hotspot centers are given in Table S1.