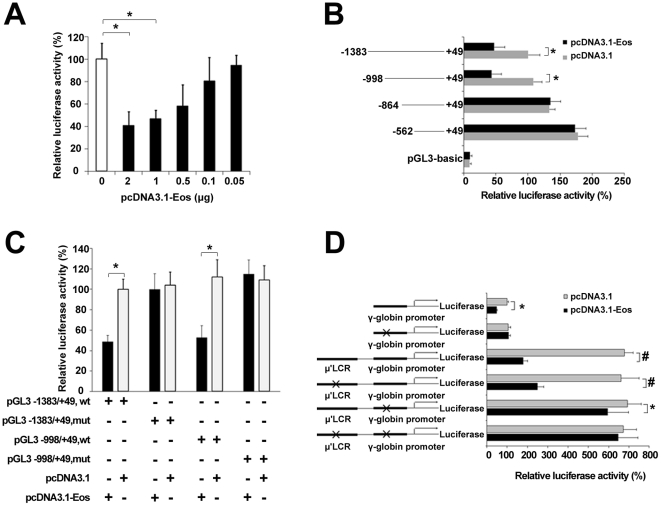
Figure 4. Validation of functional Eos-binding sites in the β-globin cluster using luciferase reporter assays.
(A) Eos represses γ-globin promoter activity in a dose-dependent manner. Luciferase reporter assays were performed in K562 cells. The recombinant pGL3-basic-1.4 kb γ-globin promoter (−1383/+49) construct was cotransfected into K562 cells with increasing amounts of the pcDNA3.1-Eos expression vector. (B) Truncation analysis of γ-globin promoter activity in K562 cells. Cells were cotransfected with pGL3-basic-γ-globin promoter constructs at various lengths and either pcDNA3.1 (i.e., empty vector) or pcDNA3.1-Eos. Promoter/reporter gene constructs with different γ-globin promoter lengths are depicted along the left. (C) Mutation analysis of γ-globin promoter activity in K562 cells. pGL3 constructs with different γ-globin promoter regions (−1383/+49 or −998/+49) and with normal or mutated sequences were cotransfected with pcDNA3.1 (i.e., empty vector) or with pcDNA3.1-Eos into K562 cells. (D) The luciferase reporter construct consists of the 3.1-kb μ'LCR, the 1.4-kb γ-globin promoter, and the luciferase reporter in the pGL3-basic plasmid (see Materials and Methods). pGL3 constructs with normal or mutated sequences in the LCR or γ-globin promoter were cotransfected with pcDNA3.1 or pcDNA3.1-Eos into K562 cells, and luciferase activities were determined. Luciferase reporter assay data are expressed as percentages of control (i.e., cells transfected with pcDNA3.1 alone) and represent the means ± SE of three separate experiments after correcting for differences in transfection efficiency by pRL-TK activities. Error bars represent one standard deviation. *P<0.05, #P<0.01.