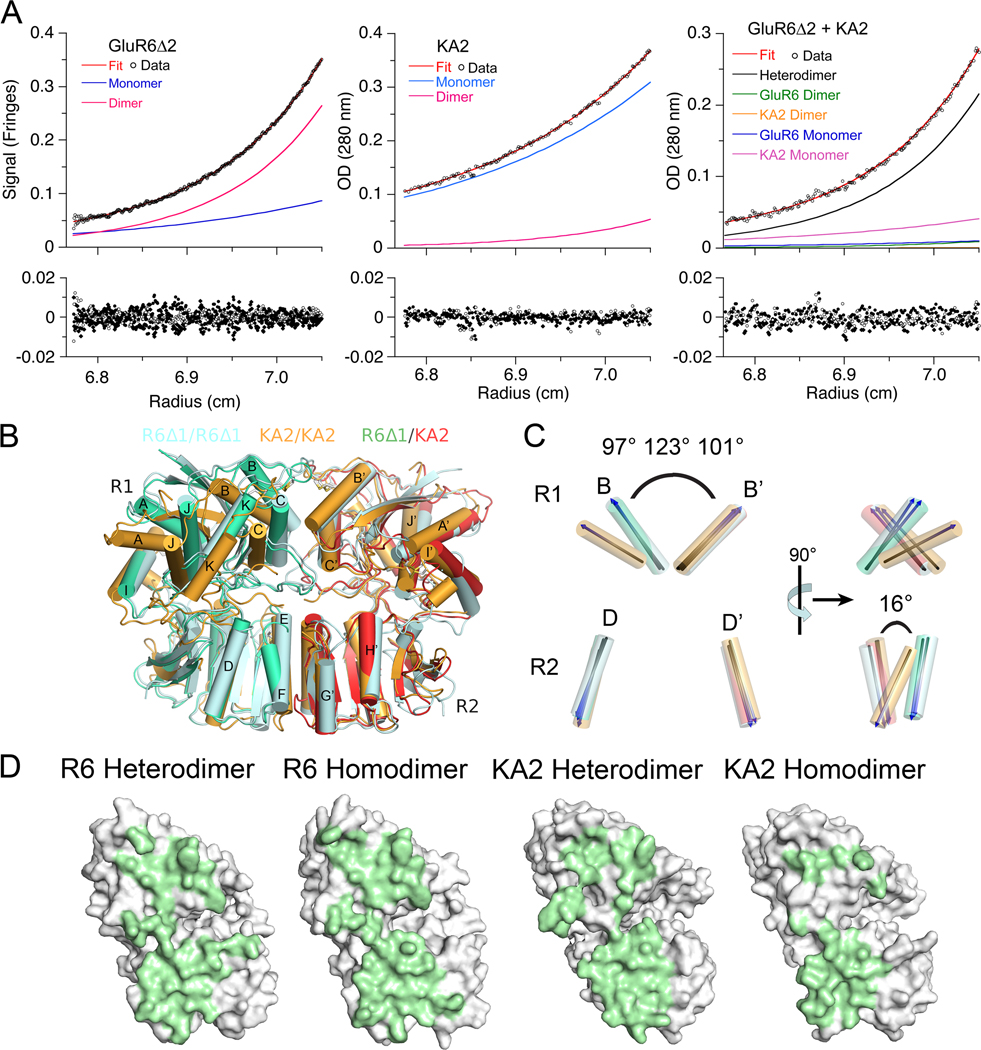
Figure 2. The GluR6 and KA2 ATDs coassemble to form high affinity heterodimers.
(A) Sedimentation equilibrium analysis for the GluR6Δ2 ATD, homodimer Kd 350 nM (left); the KA2 ATD, homodimer Kd 410 µM (middle); and an approximately equimolar mixture of the GluR6Δ2 and KA2 ATDs, heterodimer Kd 77 nM (right); fits to a monomer-dimer model (red line), with the calculated monomer and dimer populations shown as blue and pink lines, are shown for the three experiments (upper panels); the lower panels show residuals for single cells for a global fit to data for 3 loading concentrations each run at three speeds (6,500, 10,000 and 16,000 r.p.m). (B) The GluR6/KA2 heterodimer crystal structure superimposed using domain R2 coordinates on crystal structures for GluR6 (PDB 3G3H) and KA2 (PDB 3OM0) homodimers (C). Superimposed vectors drawn through helix B in domain 1 and helix D in domain 2, for the same structures, colored as in panel B, with the angle between helix B and its dimer partner indicated for the GluR6 homodimer, the KA2 homodimer, and the GluR6/KA2 heterodimer; rotation by 90° (right panel) reveals a 16° change in angle between helix D and helix D’ in domain R2 of the KA2 homodimer assembly. (D) Green shading illustrates buried molecular surfaces for the GluR6 and KA2 subunits for the same heterodimer and homodimer assemblies shown in B.