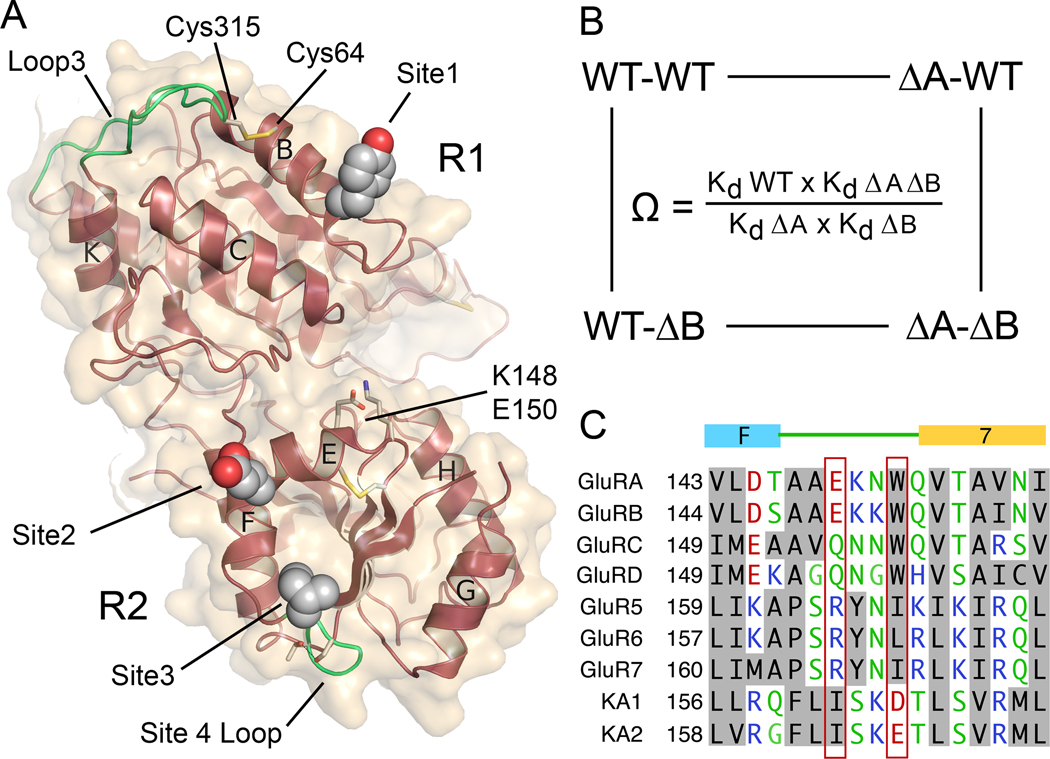
Figure 5. Mutant cycle analysis for interactions between intermolecular contacts in the heterodimer assembly.
(A) The KA2 subunit ATD viewed face on to the heterodimer surface, indicating the position of Tyr57 in site 1; Glu156 in site 2; Ile164 in site 3; Ser165 and Thr168 in site 4 that stabilizes the loop which makes site 3 contacts; the Cys64-Cys315 disulfide bond which holds loop 3 in place; and the location of Lys148 and Glu150. (B) Illustration of a mutant cycle for coupling between two sites indicated as A and B. (C) Amino acid alignment for AMPA and kainate receptors reveals a unique conservation of Ile164 in the KA2 subunits, and exchange of conserved residues in the loop connecting alpha helix F with beta strand 7.