Figure 7. Location of W68 in Kir channels.
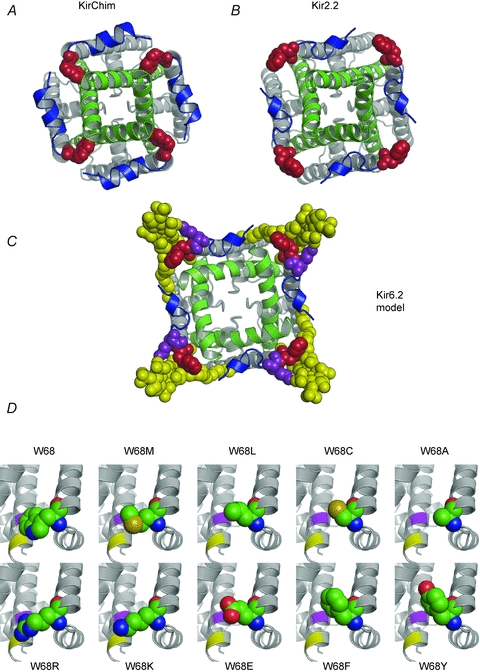
A and B, crystal structures of KirChim (PDB id: 2QKS) (A) and Kir2.2 (3JYC) (B) viewed from the intracellular side. In the majority of Kir channel X-ray structures, the residue equivalent to W68 in Kir6.2 (red) is tightly packed against the C-terminal end of TM2 (green) as seen in the crystal structure of KirChim (A). The side-chain of the residue equivalent to W68 also forms close contacts with residues in the slide helix (blue). In the Kir2.2 crystal structure (B) the tryptophan side-chain is in a different (‘flipped-out’) conformation, pointing away from the channel pore. An outward curvature of TM2 is apparent, focused on the second highly conserved glycine of TM2, although the helix–bundle crossing remains in the closed state. C, molecular model of Kir6.2 in the open state. The TM2 helices are based on KvAP; the remainder of the structure is based on Kir2.2. The ‘flipped-out’ state of W68 has allowed TM2 to undergo the opening transition. PIP2 (yellow) is shown docked into its binding site (Stansfeld et al. 2009), interacting with both W68 and with K67 (purple). D, structural models of Kir6.2 (Stansfeld et al. 2009) with the side-chain conformations of residue 68 for the mutant channels as predicted by the Hunter algorithm (Cohen et al. 2009). The locations of residues I167 (purple) and T171 (yellow) in TM2 are shown.
