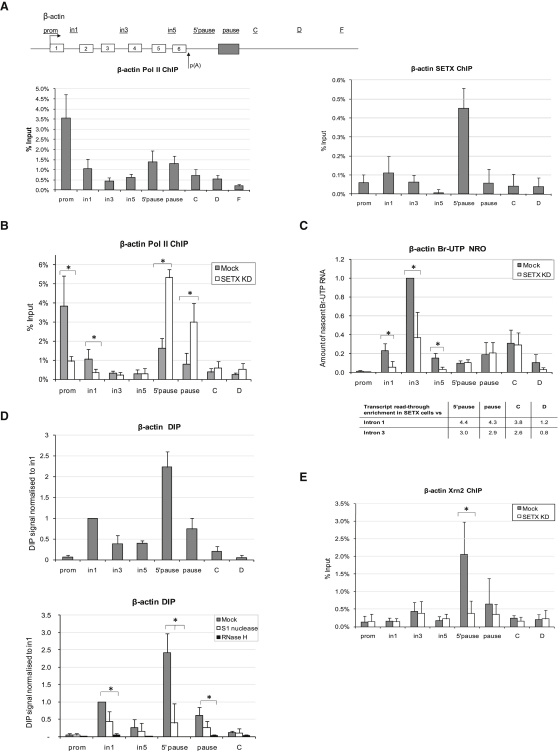
Figure 4.
Senataxin Is Required for Pol II Transcriptional Initiation, Termination, and Xrn2 Recruitment to the Termination Regions of the Endogenous β-Actin Gene
(A) Diagram of β-actin gene, with exons shown as white boxes and pause element shown as gray box. Positions of primers used in ChIP analysis are shown above the diagram. ChIP analysis was carried out with Pol II (left panel) or senataxin (right panel) antibody.
(B) Pol II ChIP analysis of β-actin gene in mock-treated (gray bars) and senataxin siRNA1-treated (white bars) cells.
(C) Br-UTP NRO analysis of β-actin gene in mock-treated (gray bars) and senataxin siRNA1-treated (white bars) cells. Amount of nascent Br-UTP RNA was normalized to intron 3 probe in mock-treated cells. All bars represent average values ± SD from at least three independent biological experiments. Enrichment of readthrough transcripts for probes “5′ pause,” “pause,” “C,” and “D” in senataxin-depleted cells, as compared to mock cells, was calculated relative to intron 1 and intron 3 signals.
(D) Top panel: DIP analysis of β-actin gene using RNA/DNA hybrids-specific S9.6 antibody, based on average values ± SD from at least three independent biological experiments. DIP signal was normalized to intron 1 signal. Bottom panel: R-loops formed over β-actin gene are sensitive to RNase H and only partially to S1 nuclease digestion. DIP samples were either untreated (gray) or treated with 10 U of RNase H (black bars) or 200 U of S1 nuclease (white bars).
(E) Xrn2 ChIP of β-actin in mock-treated (gray bars) and senataxin-depleted (white bars) HeLa cells. ChIP values are based on average values ± SD from at least three independent biological experiments. ∗ indicates statistical significance (p < 0.05), based on unpaired, two-tailed distribution Student's t-test.