Figure 4.
REV1 Is Required for Replication of G Quadruplex-Forming DNA on the Leading-Strand Template
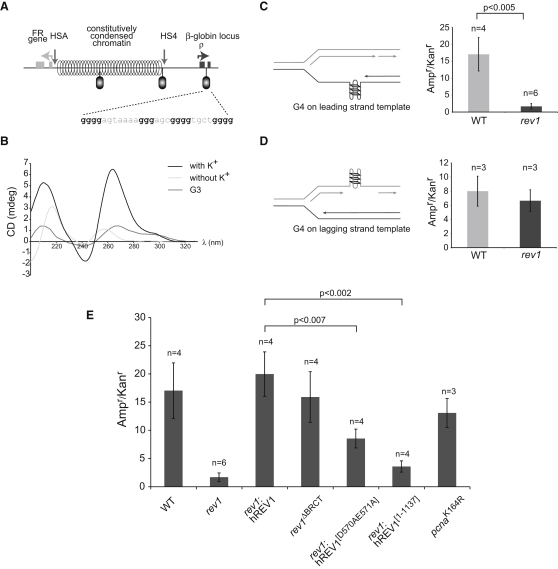
(A) Position of G4 DNAs (ovals) in the region of the β-globin locus studied in this work. The sequence of the ρ-globin G4 DNA is shown.
(B) Circular dichroism spectroscopy of the ρ-globin G quadruplex forming sequence. Renaturation of the minimal 29 bp G4 oligonucleotide (GQCDG4) in the presence (black line) or absence (light gray line) of K+ ions. Renaturation of a truncated ρ-globin G4 sequence (GQCDG3) in the presence of K+ ions (mid gray line).
(C and D)Replication efficiency, shown as the ratio of Ampr to KanrE. coli colonies, for the ρ-globin G4 DNA on the leading- (C) and lagging- (D) strand template of pQ (Szüts et al., 2008). Error bars represent standard error of the mean. p values were calculated using the unpaired t test (two-tailed).
(E) Replication efficiency of the leading-strand template G4 in rev1 mutants. Complementation is with full-length, catalytically inactive (D570AE571A) and C-terminally truncated (1-1137) human REV1. The BRCT mutant is an endogenous deletion of amino acids 69–116 of REV1 (Figure S2). The WT and rev1 data from Figure 4C are shown again for comparison. Error bars represent standard error of the mean.
See also Figures S2 and S3.